
Lactobacillus vaccinostercus DSM 20634
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Firmicutes; Bacilli; Lactobacillales; Lactobacillaceae; Paucilactobacillus; Paucilactobacillus vaccinostercus
Average proteome isoelectric point is 6.45
Get precalculated fractions of proteins
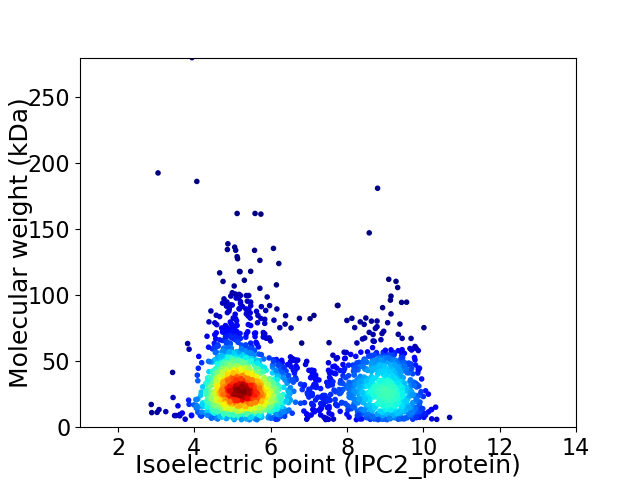
Virtual 2D-PAGE plot for 2398 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A0R2A7D5|A0A0R2A7D5_9LACO Mevalonate kinase OS=Lactobacillus vaccinostercus DSM 20634 OX=1423813 GN=FC26_GL002174 PE=4 SV=1
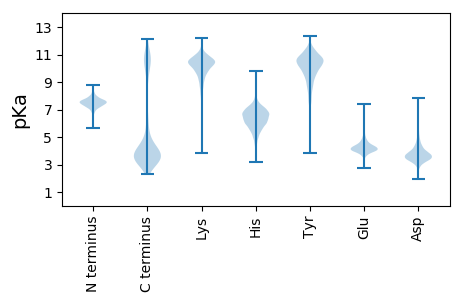
MM1 pKa = 7.54ASALVVYY8 pKa = 8.24ATITGNNEE16 pKa = 3.46DD17 pKa = 3.72VADD20 pKa = 4.59VITEE24 pKa = 3.9ALEE27 pKa = 4.18DD28 pKa = 3.81LGVDD32 pKa = 3.3VDD34 pKa = 3.96EE35 pKa = 4.97TEE37 pKa = 3.91ITQADD42 pKa = 3.56AEE44 pKa = 4.37EE45 pKa = 5.2FEE47 pKa = 5.11DD48 pKa = 5.27ADD50 pKa = 3.54ICVVCPYY57 pKa = 10.11TYY59 pKa = 10.79DD60 pKa = 3.34EE61 pKa = 4.73GALPEE66 pKa = 4.34EE67 pKa = 4.73GMDD70 pKa = 4.13FYY72 pKa = 11.74EE73 pKa = 5.79DD74 pKa = 3.77LQEE77 pKa = 5.29LDD79 pKa = 4.27LSGKK83 pKa = 9.75IYY85 pKa = 10.61GVAGSGDD92 pKa = 3.77TFYY95 pKa = 11.52LDD97 pKa = 4.59DD98 pKa = 4.08YY99 pKa = 10.62CVSVDD104 pKa = 3.22KK105 pKa = 10.68FAAAFEE111 pKa = 4.32KK112 pKa = 10.95AKK114 pKa = 9.33ATKK117 pKa = 10.01GAEE120 pKa = 3.71AVKK123 pKa = 10.6INLEE127 pKa = 4.14PEE129 pKa = 4.04NEE131 pKa = 4.94DD132 pKa = 3.74INTLNAFAKK141 pKa = 10.64SLVEE145 pKa = 4.13KK146 pKa = 11.22VNDD149 pKa = 3.57
MM1 pKa = 7.54ASALVVYY8 pKa = 8.24ATITGNNEE16 pKa = 3.46DD17 pKa = 3.72VADD20 pKa = 4.59VITEE24 pKa = 3.9ALEE27 pKa = 4.18DD28 pKa = 3.81LGVDD32 pKa = 3.3VDD34 pKa = 3.96EE35 pKa = 4.97TEE37 pKa = 3.91ITQADD42 pKa = 3.56AEE44 pKa = 4.37EE45 pKa = 5.2FEE47 pKa = 5.11DD48 pKa = 5.27ADD50 pKa = 3.54ICVVCPYY57 pKa = 10.11TYY59 pKa = 10.79DD60 pKa = 3.34EE61 pKa = 4.73GALPEE66 pKa = 4.34EE67 pKa = 4.73GMDD70 pKa = 4.13FYY72 pKa = 11.74EE73 pKa = 5.79DD74 pKa = 3.77LQEE77 pKa = 5.29LDD79 pKa = 4.27LSGKK83 pKa = 9.75IYY85 pKa = 10.61GVAGSGDD92 pKa = 3.77TFYY95 pKa = 11.52LDD97 pKa = 4.59DD98 pKa = 4.08YY99 pKa = 10.62CVSVDD104 pKa = 3.22KK105 pKa = 10.68FAAAFEE111 pKa = 4.32KK112 pKa = 10.95AKK114 pKa = 9.33ATKK117 pKa = 10.01GAEE120 pKa = 3.71AVKK123 pKa = 10.6INLEE127 pKa = 4.14PEE129 pKa = 4.04NEE131 pKa = 4.94DD132 pKa = 3.74INTLNAFAKK141 pKa = 10.64SLVEE145 pKa = 4.13KK146 pKa = 11.22VNDD149 pKa = 3.57
Molecular weight: 16.15 kDa
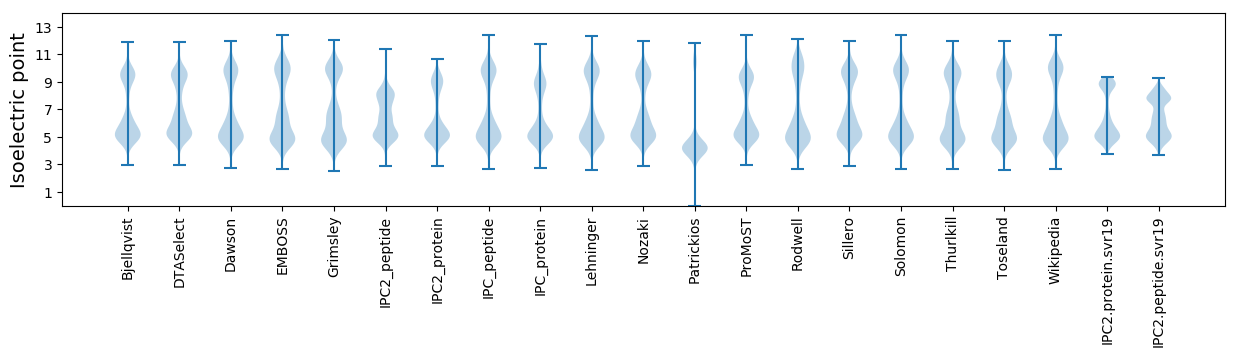
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A0R2A596|A0A0R2A596_9LACO Uncharacterized protein OS=Lactobacillus vaccinostercus DSM 20634 OX=1423813 GN=FC26_GL002472 PE=4 SV=1
MM1 pKa = 7.6RR2 pKa = 11.84RR3 pKa = 11.84IKK5 pKa = 10.31LWQLNVLLALVLISVAVLGLCTGVKK30 pKa = 9.18WEE32 pKa = 4.55SLTLLWRR39 pKa = 11.84APASLDD45 pKa = 3.34GRR47 pKa = 11.84IMWTIRR53 pKa = 11.84IPRR56 pKa = 11.84ILSVVMVGALLSVSGLLMQSLSRR79 pKa = 11.84NPIADD84 pKa = 3.74PSILGVNAGAMLALIIGSLLGMSLSIVTTIWLSLVGAAVAFTAVLLIAMTRR135 pKa = 11.84QGLDD139 pKa = 2.94VLRR142 pKa = 11.84FILGGTVFSSFISCLAYY159 pKa = 10.3AGSLLTNTTMQFRR172 pKa = 11.84NLLVGGFSAVTYY184 pKa = 9.2AHH186 pKa = 6.31VRR188 pKa = 11.84VLCGGLIIVLATILLMRR205 pKa = 11.84KK206 pKa = 8.51QLSLLVLDD214 pKa = 4.52DD215 pKa = 3.48TTARR219 pKa = 11.84SLGAKK224 pKa = 9.66PGLTRR229 pKa = 11.84VVAALLVVISAGISVAVAGNIGFVGLGIPQMINYY263 pKa = 6.97VHH265 pKa = 7.76PDD267 pKa = 2.96RR268 pKa = 11.84LEE270 pKa = 4.4RR271 pKa = 11.84NVVPTVLLGAIFMMVVDD288 pKa = 5.24IVAKK292 pKa = 9.57TLTAPSEE299 pKa = 4.3LPLSALSAICGGIFLLLVIMKK320 pKa = 8.86GARR323 pKa = 11.84VTEE326 pKa = 4.23RR327 pKa = 3.33
MM1 pKa = 7.6RR2 pKa = 11.84RR3 pKa = 11.84IKK5 pKa = 10.31LWQLNVLLALVLISVAVLGLCTGVKK30 pKa = 9.18WEE32 pKa = 4.55SLTLLWRR39 pKa = 11.84APASLDD45 pKa = 3.34GRR47 pKa = 11.84IMWTIRR53 pKa = 11.84IPRR56 pKa = 11.84ILSVVMVGALLSVSGLLMQSLSRR79 pKa = 11.84NPIADD84 pKa = 3.74PSILGVNAGAMLALIIGSLLGMSLSIVTTIWLSLVGAAVAFTAVLLIAMTRR135 pKa = 11.84QGLDD139 pKa = 2.94VLRR142 pKa = 11.84FILGGTVFSSFISCLAYY159 pKa = 10.3AGSLLTNTTMQFRR172 pKa = 11.84NLLVGGFSAVTYY184 pKa = 9.2AHH186 pKa = 6.31VRR188 pKa = 11.84VLCGGLIIVLATILLMRR205 pKa = 11.84KK206 pKa = 8.51QLSLLVLDD214 pKa = 4.52DD215 pKa = 3.48TTARR219 pKa = 11.84SLGAKK224 pKa = 9.66PGLTRR229 pKa = 11.84VVAALLVVISAGISVAVAGNIGFVGLGIPQMINYY263 pKa = 6.97VHH265 pKa = 7.76PDD267 pKa = 2.96RR268 pKa = 11.84LEE270 pKa = 4.4RR271 pKa = 11.84NVVPTVLLGAIFMMVVDD288 pKa = 5.24IVAKK292 pKa = 9.57TLTAPSEE299 pKa = 4.3LPLSALSAICGGIFLLLVIMKK320 pKa = 8.86GARR323 pKa = 11.84VTEE326 pKa = 4.23RR327 pKa = 3.33
Molecular weight: 34.56 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
735279 |
50 |
2674 |
306.6 |
34.15 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
8.282 ± 0.062 | 0.543 ± 0.013 |
5.83 ± 0.054 | 5.207 ± 0.054 |
4.214 ± 0.043 | 6.689 ± 0.045 |
2.431 ± 0.026 | 7.076 ± 0.055 |
5.516 ± 0.049 | 9.646 ± 0.069 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.763 ± 0.028 | 4.529 ± 0.037 |
3.526 ± 0.034 | 5.2 ± 0.055 |
4.155 ± 0.04 | 5.988 ± 0.111 |
6.618 ± 0.067 | 7.249 ± 0.043 |
1.045 ± 0.021 | 3.485 ± 0.034 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
