
Pepo aphid-borne yellows virus
Taxonomy: Viruses; Riboviria; Orthornavirae; Pisuviricota; Pisoniviricetes; Sobelivirales; Solemoviridae; Polerovirus
Average proteome isoelectric point is 7.15
Get precalculated fractions of proteins
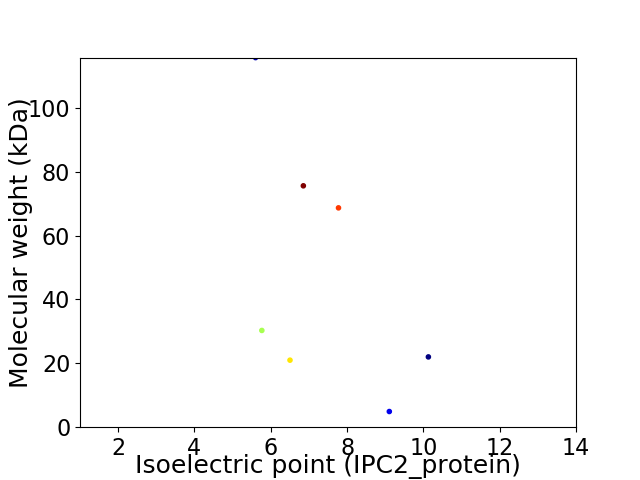
Virtual 2D-PAGE plot for 7 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A172ZQ18|A0A172ZQ18_9LUTE Readthrough protein OS=Pepo aphid-borne yellows virus OX=1462681 PE=3 SV=1
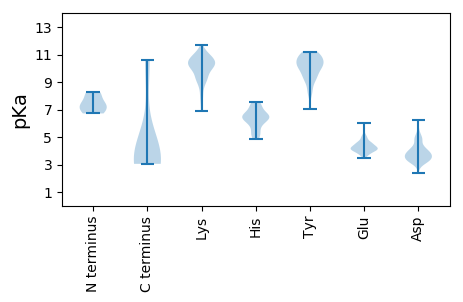
MM1 pKa = 7.35NMLFVLFFSLSLSLCGATSPVSGTALIQPQPLNFNSSFDD40 pKa = 3.82GLLSAEE46 pKa = 4.3WVSPPPEE53 pKa = 3.98VRR55 pKa = 11.84WIEE58 pKa = 4.24GTLQWPPQDD67 pKa = 4.16QEE69 pKa = 4.34EE70 pKa = 4.61TSCDD74 pKa = 3.55YY75 pKa = 10.99SYY77 pKa = 9.79HH78 pKa = 6.6TIFQKK83 pKa = 10.63LQEE86 pKa = 4.88NILEE90 pKa = 4.58DD91 pKa = 4.8GKK93 pKa = 11.32KK94 pKa = 9.92LWLGGHH100 pKa = 5.08QQLTNFSTNIYY111 pKa = 9.39NLSRR115 pKa = 11.84ALASDD120 pKa = 3.91ACTLLIDD127 pKa = 5.01SIIFLWSFILWGVVWCVFRR146 pKa = 11.84FVKK149 pKa = 10.08TYY151 pKa = 9.3FLRR154 pKa = 11.84ALTIAFLVACTACMAKK170 pKa = 10.14CLRR173 pKa = 11.84ALSGLLAPFPRR184 pKa = 11.84LLLSVINFIYY194 pKa = 10.59SVFLGMLTPFQSSKK208 pKa = 7.96TRR210 pKa = 11.84KK211 pKa = 6.9TNCWEE216 pKa = 4.06KK217 pKa = 10.49QLKK220 pKa = 8.65GWNSFEE226 pKa = 4.68IPMNPPSKK234 pKa = 10.42SVIEE238 pKa = 3.82ITYY241 pKa = 10.88DD242 pKa = 3.22NGSHH246 pKa = 6.69CGYY249 pKa = 10.65AVTVQTYY256 pKa = 10.31KK257 pKa = 9.72KK258 pKa = 7.59TASGAFEE265 pKa = 4.39SAVLTSLHH273 pKa = 6.5CLDD276 pKa = 4.94GGDD279 pKa = 3.74EE280 pKa = 4.26KK281 pKa = 11.34FLFSRR286 pKa = 11.84RR287 pKa = 11.84TGQGIPLAQCTTLAVWKK304 pKa = 10.79SLDD307 pKa = 3.72LCLLTPPANFQSTLGVKK324 pKa = 9.11PCPMVCMDD332 pKa = 5.22RR333 pKa = 11.84LAGCDD338 pKa = 3.0ASFFRR343 pKa = 11.84LNKK346 pKa = 9.17EE347 pKa = 4.56GAWSMTNCKK356 pKa = 10.14LLGAQKK362 pKa = 10.28DD363 pKa = 3.81GSVAVLVNSQPGHH376 pKa = 5.88SGSPFFNGKK385 pKa = 8.99NVLGVLRR392 pKa = 11.84GPDD395 pKa = 3.44PDD397 pKa = 3.87NNFNIMNPIPAVEE410 pKa = 4.2GLTVPEE416 pKa = 4.54LRR418 pKa = 11.84FEE420 pKa = 4.37TTAPTGKK427 pKa = 10.45LFTDD431 pKa = 4.04DD432 pKa = 4.94NLMKK436 pKa = 10.72ALAEE440 pKa = 4.2EE441 pKa = 4.29YY442 pKa = 10.24TRR444 pKa = 11.84MKK446 pKa = 10.92NFVPASGKK454 pKa = 10.0AWADD458 pKa = 3.3YY459 pKa = 11.2DD460 pKa = 3.77SDD462 pKa = 3.87EE463 pKa = 4.95EE464 pKa = 4.39YY465 pKa = 11.22FEE467 pKa = 5.6AAAKK471 pKa = 10.41DD472 pKa = 3.49LAGPSTLQGNRR483 pKa = 11.84RR484 pKa = 11.84GQARR488 pKa = 11.84LPNQRR493 pKa = 11.84GPCRR497 pKa = 11.84LPEE500 pKa = 4.26NDD502 pKa = 4.4AGNDD506 pKa = 3.67GKK508 pKa = 10.81DD509 pKa = 4.03DD510 pKa = 3.57IQHH513 pKa = 6.46FSGGHH518 pKa = 5.34KK519 pKa = 9.66RR520 pKa = 11.84DD521 pKa = 3.8CGGEE525 pKa = 3.91AGRR528 pKa = 11.84KK529 pKa = 7.93AVPYY533 pKa = 8.63EE534 pKa = 3.74QASPEE539 pKa = 4.06EE540 pKa = 4.16EE541 pKa = 4.41PPVSSAKK548 pKa = 9.43QAEE551 pKa = 4.48DD552 pKa = 3.31FRR554 pKa = 11.84QYY556 pKa = 11.22FNSLYY561 pKa = 10.26KK562 pKa = 9.82WDD564 pKa = 3.53VCAPSSEE571 pKa = 4.25VPGFRR576 pKa = 11.84NCGHH580 pKa = 7.55LPQFYY585 pKa = 10.39RR586 pKa = 11.84PGKK589 pKa = 9.28KK590 pKa = 10.06AEE592 pKa = 4.25TEE594 pKa = 3.72WGKK597 pKa = 7.81TLTKK601 pKa = 10.07EE602 pKa = 4.09YY603 pKa = 9.06PALGAATRR611 pKa = 11.84GFGWPEE617 pKa = 3.79FGPAAEE623 pKa = 4.51LKK625 pKa = 10.44SLRR628 pKa = 11.84LQSARR633 pKa = 11.84WRR635 pKa = 11.84EE636 pKa = 3.97RR637 pKa = 11.84ASSATEE643 pKa = 3.69PCQSEE648 pKa = 4.49RR649 pKa = 11.84EE650 pKa = 4.06RR651 pKa = 11.84VIAEE655 pKa = 3.61LTEE658 pKa = 4.43RR659 pKa = 11.84YY660 pKa = 10.52SNVKK664 pKa = 9.96SCAPACARR672 pKa = 11.84QEE674 pKa = 4.17TLSWDD679 pKa = 3.61GFLEE683 pKa = 4.83DD684 pKa = 4.68IKK686 pKa = 11.28SAIPFLSLDD695 pKa = 3.51AGVGVPYY702 pKa = 10.2IAYY705 pKa = 8.32GKK707 pKa = 7.16PTLAQWVHH715 pKa = 6.69DD716 pKa = 4.41EE717 pKa = 4.23SLLPLVARR725 pKa = 11.84LAFDD729 pKa = 3.64RR730 pKa = 11.84LKK732 pKa = 11.17KK733 pKa = 9.25MSEE736 pKa = 4.02VSFEE740 pKa = 4.04ALSPEE745 pKa = 4.1EE746 pKa = 4.04LVQEE750 pKa = 4.59GLCDD754 pKa = 4.87PIRR757 pKa = 11.84VFVKK761 pKa = 10.78GEE763 pKa = 3.59PHH765 pKa = 6.19KK766 pKa = 10.54QSKK769 pKa = 10.21LDD771 pKa = 3.38EE772 pKa = 4.27GRR774 pKa = 11.84YY775 pKa = 8.96RR776 pKa = 11.84LIMSVSMVDD785 pKa = 3.26QLVARR790 pKa = 11.84VLFQNQNRR798 pKa = 11.84AEE800 pKa = 4.07IALWRR805 pKa = 11.84AIPSKK810 pKa = 10.48PGLGLSTDD818 pKa = 3.54EE819 pKa = 4.94QITDD823 pKa = 4.25FAQNLSQYY831 pKa = 9.23VRR833 pKa = 11.84EE834 pKa = 4.14SRR836 pKa = 11.84GDD838 pKa = 3.31IIEE841 pKa = 4.3RR842 pKa = 11.84WHH844 pKa = 6.26EE845 pKa = 4.1CVVPTDD851 pKa = 3.67CSGFDD856 pKa = 3.34WSVSSWMLEE865 pKa = 3.63DD866 pKa = 6.11DD867 pKa = 3.23IVVRR871 pKa = 11.84NNLTLDD877 pKa = 3.27NTEE880 pKa = 3.63LTKK883 pKa = 10.56RR884 pKa = 11.84LRR886 pKa = 11.84AAWLKK891 pKa = 10.89CIANSVLCLSDD902 pKa = 3.35GTLLAQEE909 pKa = 4.44FPGVQKK915 pKa = 10.71SGSYY919 pKa = 8.25NTSSSNSRR927 pKa = 11.84IRR929 pKa = 11.84VMASIYY935 pKa = 10.79AGASWCIAMGDD946 pKa = 3.91DD947 pKa = 4.33ALEE950 pKa = 4.7SSDD953 pKa = 4.02TDD955 pKa = 3.64LKK957 pKa = 11.25VYY959 pKa = 10.84SDD961 pKa = 4.52LGLKK965 pKa = 10.27VEE967 pKa = 4.53VSDD970 pKa = 3.87KK971 pKa = 11.68LEE973 pKa = 4.33FCSHH977 pKa = 6.49HH978 pKa = 6.35FKK980 pKa = 10.84SSSLAIPVNVGKK992 pKa = 9.06MLYY995 pKa = 10.42KK996 pKa = 10.55LIHH999 pKa = 6.81GYY1001 pKa = 9.62NIRR1004 pKa = 11.84PGCTSVEE1011 pKa = 4.23VIANYY1016 pKa = 9.49MSACYY1021 pKa = 9.94SVLNEE1026 pKa = 4.0LRR1028 pKa = 11.84HH1029 pKa = 5.31VPEE1032 pKa = 5.62AIPLVSACLFSSS1044 pKa = 3.92
MM1 pKa = 7.35NMLFVLFFSLSLSLCGATSPVSGTALIQPQPLNFNSSFDD40 pKa = 3.82GLLSAEE46 pKa = 4.3WVSPPPEE53 pKa = 3.98VRR55 pKa = 11.84WIEE58 pKa = 4.24GTLQWPPQDD67 pKa = 4.16QEE69 pKa = 4.34EE70 pKa = 4.61TSCDD74 pKa = 3.55YY75 pKa = 10.99SYY77 pKa = 9.79HH78 pKa = 6.6TIFQKK83 pKa = 10.63LQEE86 pKa = 4.88NILEE90 pKa = 4.58DD91 pKa = 4.8GKK93 pKa = 11.32KK94 pKa = 9.92LWLGGHH100 pKa = 5.08QQLTNFSTNIYY111 pKa = 9.39NLSRR115 pKa = 11.84ALASDD120 pKa = 3.91ACTLLIDD127 pKa = 5.01SIIFLWSFILWGVVWCVFRR146 pKa = 11.84FVKK149 pKa = 10.08TYY151 pKa = 9.3FLRR154 pKa = 11.84ALTIAFLVACTACMAKK170 pKa = 10.14CLRR173 pKa = 11.84ALSGLLAPFPRR184 pKa = 11.84LLLSVINFIYY194 pKa = 10.59SVFLGMLTPFQSSKK208 pKa = 7.96TRR210 pKa = 11.84KK211 pKa = 6.9TNCWEE216 pKa = 4.06KK217 pKa = 10.49QLKK220 pKa = 8.65GWNSFEE226 pKa = 4.68IPMNPPSKK234 pKa = 10.42SVIEE238 pKa = 3.82ITYY241 pKa = 10.88DD242 pKa = 3.22NGSHH246 pKa = 6.69CGYY249 pKa = 10.65AVTVQTYY256 pKa = 10.31KK257 pKa = 9.72KK258 pKa = 7.59TASGAFEE265 pKa = 4.39SAVLTSLHH273 pKa = 6.5CLDD276 pKa = 4.94GGDD279 pKa = 3.74EE280 pKa = 4.26KK281 pKa = 11.34FLFSRR286 pKa = 11.84RR287 pKa = 11.84TGQGIPLAQCTTLAVWKK304 pKa = 10.79SLDD307 pKa = 3.72LCLLTPPANFQSTLGVKK324 pKa = 9.11PCPMVCMDD332 pKa = 5.22RR333 pKa = 11.84LAGCDD338 pKa = 3.0ASFFRR343 pKa = 11.84LNKK346 pKa = 9.17EE347 pKa = 4.56GAWSMTNCKK356 pKa = 10.14LLGAQKK362 pKa = 10.28DD363 pKa = 3.81GSVAVLVNSQPGHH376 pKa = 5.88SGSPFFNGKK385 pKa = 8.99NVLGVLRR392 pKa = 11.84GPDD395 pKa = 3.44PDD397 pKa = 3.87NNFNIMNPIPAVEE410 pKa = 4.2GLTVPEE416 pKa = 4.54LRR418 pKa = 11.84FEE420 pKa = 4.37TTAPTGKK427 pKa = 10.45LFTDD431 pKa = 4.04DD432 pKa = 4.94NLMKK436 pKa = 10.72ALAEE440 pKa = 4.2EE441 pKa = 4.29YY442 pKa = 10.24TRR444 pKa = 11.84MKK446 pKa = 10.92NFVPASGKK454 pKa = 10.0AWADD458 pKa = 3.3YY459 pKa = 11.2DD460 pKa = 3.77SDD462 pKa = 3.87EE463 pKa = 4.95EE464 pKa = 4.39YY465 pKa = 11.22FEE467 pKa = 5.6AAAKK471 pKa = 10.41DD472 pKa = 3.49LAGPSTLQGNRR483 pKa = 11.84RR484 pKa = 11.84GQARR488 pKa = 11.84LPNQRR493 pKa = 11.84GPCRR497 pKa = 11.84LPEE500 pKa = 4.26NDD502 pKa = 4.4AGNDD506 pKa = 3.67GKK508 pKa = 10.81DD509 pKa = 4.03DD510 pKa = 3.57IQHH513 pKa = 6.46FSGGHH518 pKa = 5.34KK519 pKa = 9.66RR520 pKa = 11.84DD521 pKa = 3.8CGGEE525 pKa = 3.91AGRR528 pKa = 11.84KK529 pKa = 7.93AVPYY533 pKa = 8.63EE534 pKa = 3.74QASPEE539 pKa = 4.06EE540 pKa = 4.16EE541 pKa = 4.41PPVSSAKK548 pKa = 9.43QAEE551 pKa = 4.48DD552 pKa = 3.31FRR554 pKa = 11.84QYY556 pKa = 11.22FNSLYY561 pKa = 10.26KK562 pKa = 9.82WDD564 pKa = 3.53VCAPSSEE571 pKa = 4.25VPGFRR576 pKa = 11.84NCGHH580 pKa = 7.55LPQFYY585 pKa = 10.39RR586 pKa = 11.84PGKK589 pKa = 9.28KK590 pKa = 10.06AEE592 pKa = 4.25TEE594 pKa = 3.72WGKK597 pKa = 7.81TLTKK601 pKa = 10.07EE602 pKa = 4.09YY603 pKa = 9.06PALGAATRR611 pKa = 11.84GFGWPEE617 pKa = 3.79FGPAAEE623 pKa = 4.51LKK625 pKa = 10.44SLRR628 pKa = 11.84LQSARR633 pKa = 11.84WRR635 pKa = 11.84EE636 pKa = 3.97RR637 pKa = 11.84ASSATEE643 pKa = 3.69PCQSEE648 pKa = 4.49RR649 pKa = 11.84EE650 pKa = 4.06RR651 pKa = 11.84VIAEE655 pKa = 3.61LTEE658 pKa = 4.43RR659 pKa = 11.84YY660 pKa = 10.52SNVKK664 pKa = 9.96SCAPACARR672 pKa = 11.84QEE674 pKa = 4.17TLSWDD679 pKa = 3.61GFLEE683 pKa = 4.83DD684 pKa = 4.68IKK686 pKa = 11.28SAIPFLSLDD695 pKa = 3.51AGVGVPYY702 pKa = 10.2IAYY705 pKa = 8.32GKK707 pKa = 7.16PTLAQWVHH715 pKa = 6.69DD716 pKa = 4.41EE717 pKa = 4.23SLLPLVARR725 pKa = 11.84LAFDD729 pKa = 3.64RR730 pKa = 11.84LKK732 pKa = 11.17KK733 pKa = 9.25MSEE736 pKa = 4.02VSFEE740 pKa = 4.04ALSPEE745 pKa = 4.1EE746 pKa = 4.04LVQEE750 pKa = 4.59GLCDD754 pKa = 4.87PIRR757 pKa = 11.84VFVKK761 pKa = 10.78GEE763 pKa = 3.59PHH765 pKa = 6.19KK766 pKa = 10.54QSKK769 pKa = 10.21LDD771 pKa = 3.38EE772 pKa = 4.27GRR774 pKa = 11.84YY775 pKa = 8.96RR776 pKa = 11.84LIMSVSMVDD785 pKa = 3.26QLVARR790 pKa = 11.84VLFQNQNRR798 pKa = 11.84AEE800 pKa = 4.07IALWRR805 pKa = 11.84AIPSKK810 pKa = 10.48PGLGLSTDD818 pKa = 3.54EE819 pKa = 4.94QITDD823 pKa = 4.25FAQNLSQYY831 pKa = 9.23VRR833 pKa = 11.84EE834 pKa = 4.14SRR836 pKa = 11.84GDD838 pKa = 3.31IIEE841 pKa = 4.3RR842 pKa = 11.84WHH844 pKa = 6.26EE845 pKa = 4.1CVVPTDD851 pKa = 3.67CSGFDD856 pKa = 3.34WSVSSWMLEE865 pKa = 3.63DD866 pKa = 6.11DD867 pKa = 3.23IVVRR871 pKa = 11.84NNLTLDD877 pKa = 3.27NTEE880 pKa = 3.63LTKK883 pKa = 10.56RR884 pKa = 11.84LRR886 pKa = 11.84AAWLKK891 pKa = 10.89CIANSVLCLSDD902 pKa = 3.35GTLLAQEE909 pKa = 4.44FPGVQKK915 pKa = 10.71SGSYY919 pKa = 8.25NTSSSNSRR927 pKa = 11.84IRR929 pKa = 11.84VMASIYY935 pKa = 10.79AGASWCIAMGDD946 pKa = 3.91DD947 pKa = 4.33ALEE950 pKa = 4.7SSDD953 pKa = 4.02TDD955 pKa = 3.64LKK957 pKa = 11.25VYY959 pKa = 10.84SDD961 pKa = 4.52LGLKK965 pKa = 10.27VEE967 pKa = 4.53VSDD970 pKa = 3.87KK971 pKa = 11.68LEE973 pKa = 4.33FCSHH977 pKa = 6.49HH978 pKa = 6.35FKK980 pKa = 10.84SSSLAIPVNVGKK992 pKa = 9.06MLYY995 pKa = 10.42KK996 pKa = 10.55LIHH999 pKa = 6.81GYY1001 pKa = 9.62NIRR1004 pKa = 11.84PGCTSVEE1011 pKa = 4.23VIANYY1016 pKa = 9.49MSACYY1021 pKa = 9.94SVLNEE1026 pKa = 4.0LRR1028 pKa = 11.84HH1029 pKa = 5.31VPEE1032 pKa = 5.62AIPLVSACLFSSS1044 pKa = 3.92
Molecular weight: 115.73 kDa
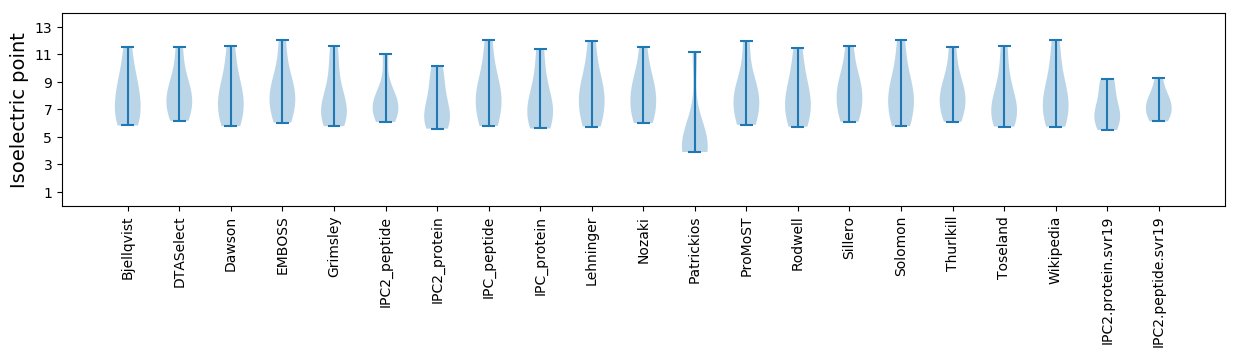
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A172ZPW1|A0A172ZPW1_9LUTE Serine protease OS=Pepo aphid-borne yellows virus OX=1462681 PE=4 SV=1
MM1 pKa = 6.72NTVAVRR7 pKa = 11.84SSNGTRR13 pKa = 11.84RR14 pKa = 11.84RR15 pKa = 11.84RR16 pKa = 11.84RR17 pKa = 11.84PRR19 pKa = 11.84RR20 pKa = 11.84SARR23 pKa = 11.84RR24 pKa = 11.84VRR26 pKa = 11.84VAMVQAIGPAQRR38 pKa = 11.84GGRR41 pKa = 11.84RR42 pKa = 11.84RR43 pKa = 11.84RR44 pKa = 11.84NRR46 pKa = 11.84RR47 pKa = 11.84RR48 pKa = 11.84VNRR51 pKa = 11.84GGRR54 pKa = 11.84ARR56 pKa = 11.84GGRR59 pKa = 11.84QGEE62 pKa = 4.26TFVFSKK68 pKa = 11.12DD69 pKa = 3.51NLSGSSSGSITFGPSLSEE87 pKa = 4.04SPAFSSGILKK97 pKa = 10.26AYY99 pKa = 10.13HH100 pKa = 6.54EE101 pKa = 4.61YY102 pKa = 10.53KK103 pKa = 9.31ITMVKK108 pKa = 10.64LEE110 pKa = 4.9FISEE114 pKa = 4.18ASSTSSGSISYY125 pKa = 10.24EE126 pKa = 3.75LDD128 pKa = 3.23PHH130 pKa = 6.86CKK132 pKa = 10.08LSSLQSTVNKK142 pKa = 10.31FGITKK147 pKa = 10.11NGSRR151 pKa = 11.84SWTAKK156 pKa = 10.17FINGLEE162 pKa = 4.02WHH164 pKa = 7.33DD165 pKa = 3.56ATEE168 pKa = 4.09DD169 pKa = 3.41QFRR172 pKa = 11.84ILYY175 pKa = 9.14KK176 pKa = 10.93GNGSSSVAGSFKK188 pKa = 9.88ITIQCQVQNPKK199 pKa = 10.62
MM1 pKa = 6.72NTVAVRR7 pKa = 11.84SSNGTRR13 pKa = 11.84RR14 pKa = 11.84RR15 pKa = 11.84RR16 pKa = 11.84RR17 pKa = 11.84PRR19 pKa = 11.84RR20 pKa = 11.84SARR23 pKa = 11.84RR24 pKa = 11.84VRR26 pKa = 11.84VAMVQAIGPAQRR38 pKa = 11.84GGRR41 pKa = 11.84RR42 pKa = 11.84RR43 pKa = 11.84RR44 pKa = 11.84NRR46 pKa = 11.84RR47 pKa = 11.84RR48 pKa = 11.84VNRR51 pKa = 11.84GGRR54 pKa = 11.84ARR56 pKa = 11.84GGRR59 pKa = 11.84QGEE62 pKa = 4.26TFVFSKK68 pKa = 11.12DD69 pKa = 3.51NLSGSSSGSITFGPSLSEE87 pKa = 4.04SPAFSSGILKK97 pKa = 10.26AYY99 pKa = 10.13HH100 pKa = 6.54EE101 pKa = 4.61YY102 pKa = 10.53KK103 pKa = 9.31ITMVKK108 pKa = 10.64LEE110 pKa = 4.9FISEE114 pKa = 4.18ASSTSSGSISYY125 pKa = 10.24EE126 pKa = 3.75LDD128 pKa = 3.23PHH130 pKa = 6.86CKK132 pKa = 10.08LSSLQSTVNKK142 pKa = 10.31FGITKK147 pKa = 10.11NGSRR151 pKa = 11.84SWTAKK156 pKa = 10.17FINGLEE162 pKa = 4.02WHH164 pKa = 7.33DD165 pKa = 3.56ATEE168 pKa = 4.09DD169 pKa = 3.41QFRR172 pKa = 11.84ILYY175 pKa = 9.14KK176 pKa = 10.93GNGSSSVAGSFKK188 pKa = 9.88ITIQCQVQNPKK199 pKa = 10.62
Molecular weight: 22.01 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
3048 |
45 |
1044 |
435.4 |
48.31 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.185 ± 0.443 | 2.165 ± 0.549 |
4.724 ± 0.546 | 5.971 ± 0.382 |
4.462 ± 0.648 | 7.415 ± 0.544 |
1.739 ± 0.321 | 4.331 ± 0.321 |
4.954 ± 0.548 | 8.957 ± 1.174 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.739 ± 0.297 | 4.626 ± 0.295 |
6.299 ± 0.463 | 3.707 ± 0.193 |
6.398 ± 1.067 | 10.039 ± 0.974 |
5.184 ± 0.385 | 5.381 ± 0.455 |
2.034 ± 0.183 | 2.657 ± 0.259 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
