
Achromatium sp. WMS1
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Gammaproteobacteria; Thiotrichales; Thiotrichaceae; Achromatium; unclassified Achromatium
Average proteome isoelectric point is 6.75
Get precalculated fractions of proteins
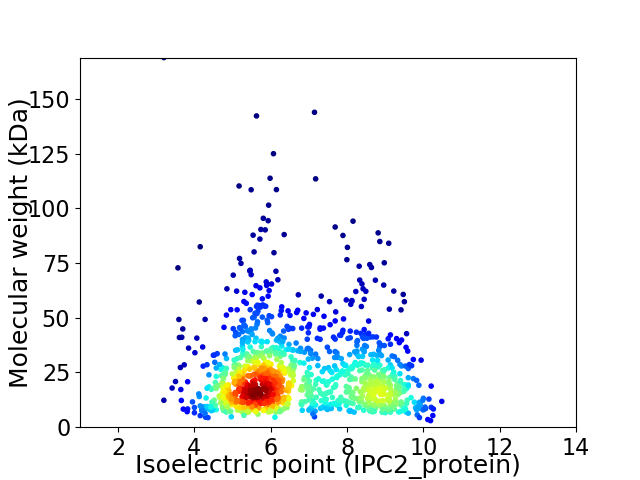
Virtual 2D-PAGE plot for 1173 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A0M1J3N3|A0A0M1J3N3_9GAMM Uncharacterized protein (Fragment) OS=Achromatium sp. WMS1 OX=1604834 GN=TI03_07095 PE=4 SV=1
RR1 pKa = 8.48VVVLATDD8 pKa = 3.71YY9 pKa = 11.41DD10 pKa = 3.72FHH12 pKa = 7.44QAGDD16 pKa = 3.86HH17 pKa = 6.46SSVMPNNGDD26 pKa = 3.52TVLDD30 pKa = 4.09GDD32 pKa = 4.27PPGTGEE38 pKa = 5.23DD39 pKa = 4.59YY40 pKa = 8.84PTIDD44 pKa = 3.24QVKK47 pKa = 9.98DD48 pKa = 3.34ALVEE52 pKa = 4.04ANIYY56 pKa = 10.09PIFAVTYY63 pKa = 8.57GVQSSYY69 pKa = 11.84DD70 pKa = 3.51DD71 pKa = 4.14LVNQLGRR78 pKa = 11.84GDD80 pKa = 3.98TVEE83 pKa = 4.58LSSDD87 pKa = 3.06SSNIVSSIQTGLEE100 pKa = 3.71NYY102 pKa = 9.84KK103 pKa = 10.72ADD105 pKa = 4.53FIEE108 pKa = 4.5NVISTPYY115 pKa = 11.01DD116 pKa = 3.22DD117 pKa = 4.55TLTGNSLDD125 pKa = 3.65NRR127 pKa = 11.84IEE129 pKa = 4.23GGAGEE134 pKa = 4.6DD135 pKa = 3.64TLKK138 pKa = 11.29GLGGDD143 pKa = 3.76DD144 pKa = 3.75TLVGGDD150 pKa = 4.19DD151 pKa = 4.57ADD153 pKa = 3.79TAIYY157 pKa = 9.86RR158 pKa = 11.84GVYY161 pKa = 8.47STVPGEE167 pKa = 3.95TSEE170 pKa = 4.65YY171 pKa = 9.59EE172 pKa = 4.15VTRR175 pKa = 11.84TRR177 pKa = 11.84LDD179 pKa = 3.31VTVKK183 pKa = 10.85DD184 pKa = 4.1LVDD187 pKa = 3.73NRR189 pKa = 11.84DD190 pKa = 3.69DD191 pKa = 3.94TDD193 pKa = 3.47TLTDD197 pKa = 3.31IEE199 pKa = 4.55TLQFANGSIDD209 pKa = 3.73VSSLPVEE216 pKa = 4.4DD217 pKa = 4.91PSINFYY223 pKa = 10.2WSARR227 pKa = 11.84TIASIINNCYY237 pKa = 10.08QVMAFNWMNAICYY250 pKa = 8.6AAA252 pKa = 4.6
RR1 pKa = 8.48VVVLATDD8 pKa = 3.71YY9 pKa = 11.41DD10 pKa = 3.72FHH12 pKa = 7.44QAGDD16 pKa = 3.86HH17 pKa = 6.46SSVMPNNGDD26 pKa = 3.52TVLDD30 pKa = 4.09GDD32 pKa = 4.27PPGTGEE38 pKa = 5.23DD39 pKa = 4.59YY40 pKa = 8.84PTIDD44 pKa = 3.24QVKK47 pKa = 9.98DD48 pKa = 3.34ALVEE52 pKa = 4.04ANIYY56 pKa = 10.09PIFAVTYY63 pKa = 8.57GVQSSYY69 pKa = 11.84DD70 pKa = 3.51DD71 pKa = 4.14LVNQLGRR78 pKa = 11.84GDD80 pKa = 3.98TVEE83 pKa = 4.58LSSDD87 pKa = 3.06SSNIVSSIQTGLEE100 pKa = 3.71NYY102 pKa = 9.84KK103 pKa = 10.72ADD105 pKa = 4.53FIEE108 pKa = 4.5NVISTPYY115 pKa = 11.01DD116 pKa = 3.22DD117 pKa = 4.55TLTGNSLDD125 pKa = 3.65NRR127 pKa = 11.84IEE129 pKa = 4.23GGAGEE134 pKa = 4.6DD135 pKa = 3.64TLKK138 pKa = 11.29GLGGDD143 pKa = 3.76DD144 pKa = 3.75TLVGGDD150 pKa = 4.19DD151 pKa = 4.57ADD153 pKa = 3.79TAIYY157 pKa = 9.86RR158 pKa = 11.84GVYY161 pKa = 8.47STVPGEE167 pKa = 3.95TSEE170 pKa = 4.65YY171 pKa = 9.59EE172 pKa = 4.15VTRR175 pKa = 11.84TRR177 pKa = 11.84LDD179 pKa = 3.31VTVKK183 pKa = 10.85DD184 pKa = 4.1LVDD187 pKa = 3.73NRR189 pKa = 11.84DD190 pKa = 3.69DD191 pKa = 3.94TDD193 pKa = 3.47TLTDD197 pKa = 3.31IEE199 pKa = 4.55TLQFANGSIDD209 pKa = 3.73VSSLPVEE216 pKa = 4.4DD217 pKa = 4.91PSINFYY223 pKa = 10.2WSARR227 pKa = 11.84TIASIINNCYY237 pKa = 10.08QVMAFNWMNAICYY250 pKa = 8.6AAA252 pKa = 4.6
Molecular weight: 27.28 kDa
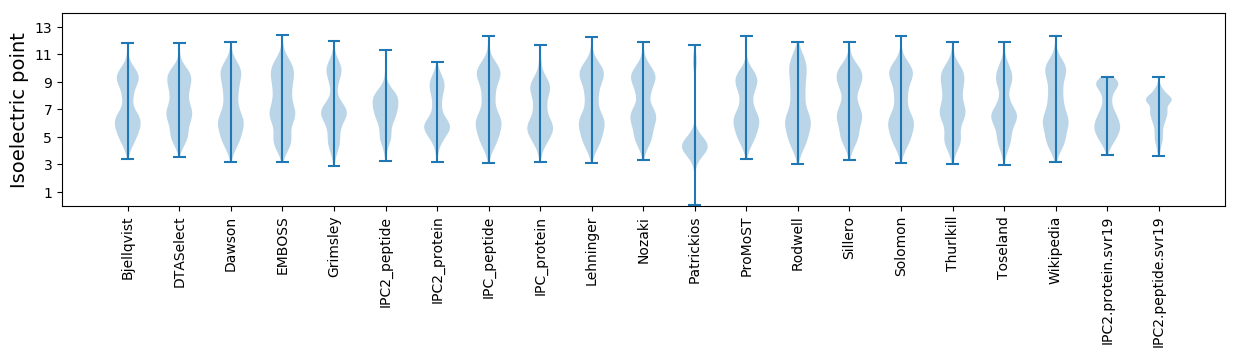
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A0M1J9Z0|A0A0M1J9Z0_9GAMM Nitric oxide reductase OS=Achromatium sp. WMS1 OX=1604834 GN=TI03_01640 PE=4 SV=1
MM1 pKa = 7.69PSFWQLRR8 pKa = 11.84CASYY12 pKa = 10.28IVKK15 pKa = 10.45AGGIIAYY22 pKa = 6.58PTEE25 pKa = 3.95AVFGLGCDD33 pKa = 4.09PYY35 pKa = 11.21NQRR38 pKa = 11.84ALTKK42 pKa = 10.04ILALKK47 pKa = 9.62GRR49 pKa = 11.84PINKK53 pKa = 9.4GVILIAADD61 pKa = 4.24FNQIIPFIAPLSNLPAARR79 pKa = 11.84ITQIKK84 pKa = 9.71ASWPGPVTWVLPARR98 pKa = 11.84NDD100 pKa = 3.22IPHH103 pKa = 7.28WLTGGRR109 pKa = 11.84HH110 pKa = 5.56TIAVRR115 pKa = 11.84ITAHH119 pKa = 6.53PSAVALCRR127 pKa = 11.84ICKK130 pKa = 7.87TALVSTSANSTNHH143 pKa = 5.88LPLRR147 pKa = 11.84NSLQLRR153 pKa = 11.84RR154 pKa = 11.84WLQRR158 pKa = 11.84LPGNNEE164 pKa = 3.18NHH166 pKa = 6.53INYY169 pKa = 9.32ILPGYY174 pKa = 9.16CGPTSRR180 pKa = 11.84PSRR183 pKa = 11.84IIDD186 pKa = 3.45ATSGLTLRR194 pKa = 11.84HH195 pKa = 5.73
MM1 pKa = 7.69PSFWQLRR8 pKa = 11.84CASYY12 pKa = 10.28IVKK15 pKa = 10.45AGGIIAYY22 pKa = 6.58PTEE25 pKa = 3.95AVFGLGCDD33 pKa = 4.09PYY35 pKa = 11.21NQRR38 pKa = 11.84ALTKK42 pKa = 10.04ILALKK47 pKa = 9.62GRR49 pKa = 11.84PINKK53 pKa = 9.4GVILIAADD61 pKa = 4.24FNQIIPFIAPLSNLPAARR79 pKa = 11.84ITQIKK84 pKa = 9.71ASWPGPVTWVLPARR98 pKa = 11.84NDD100 pKa = 3.22IPHH103 pKa = 7.28WLTGGRR109 pKa = 11.84HH110 pKa = 5.56TIAVRR115 pKa = 11.84ITAHH119 pKa = 6.53PSAVALCRR127 pKa = 11.84ICKK130 pKa = 7.87TALVSTSANSTNHH143 pKa = 5.88LPLRR147 pKa = 11.84NSLQLRR153 pKa = 11.84RR154 pKa = 11.84WLQRR158 pKa = 11.84LPGNNEE164 pKa = 3.18NHH166 pKa = 6.53INYY169 pKa = 9.32ILPGYY174 pKa = 9.16CGPTSRR180 pKa = 11.84PSRR183 pKa = 11.84IIDD186 pKa = 3.45ATSGLTLRR194 pKa = 11.84HH195 pKa = 5.73
Molecular weight: 21.4 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
269738 |
28 |
1634 |
230.0 |
25.69 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
8.219 ± 0.087 | 1.096 ± 0.032 |
5.133 ± 0.079 | 5.458 ± 0.068 |
3.786 ± 0.044 | 6.473 ± 0.107 |
2.473 ± 0.047 | 7.596 ± 0.071 |
5.055 ± 0.07 | 10.742 ± 0.088 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.248 ± 0.038 | 4.784 ± 0.068 |
4.373 ± 0.057 | 4.91 ± 0.085 |
5.087 ± 0.066 | 5.69 ± 0.056 |
6.282 ± 0.074 | 6.185 ± 0.07 |
1.315 ± 0.031 | 3.097 ± 0.044 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
