
Phalaenopsis equestris amalgavirus 1
Taxonomy: Viruses; Riboviria; Orthornavirae; Pisuviricota; Duplopiviricetes; Durnavirales; Amalgaviridae; unclassified Amalgaviridae
Average proteome isoelectric point is 6.38
Get precalculated fractions of proteins
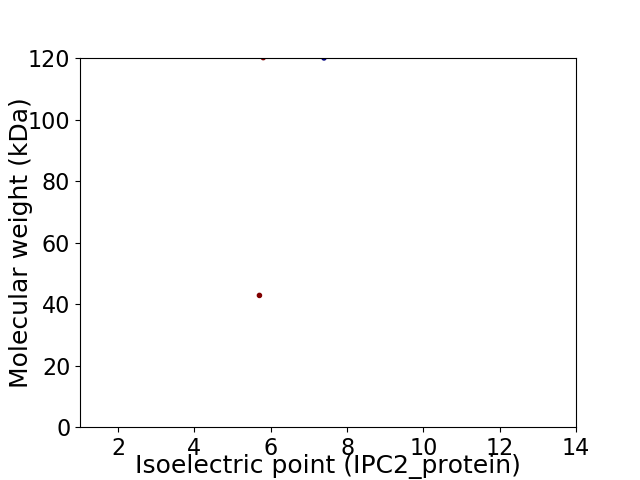
Virtual 2D-PAGE plot for 2 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A3G9M863|A0A3G9M863_9VIRU ORF1+2p OS=Phalaenopsis equestris amalgavirus 1 OX=2069327 GN=ORF1+ORF2 PE=4 SV=1
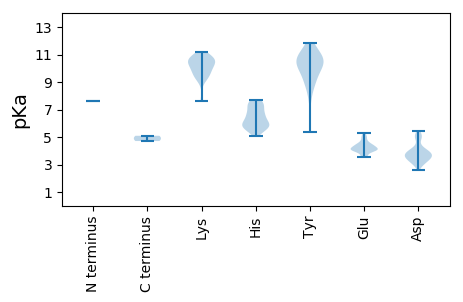
MM1 pKa = 7.6SGPSQRR7 pKa = 11.84VVHH10 pKa = 6.16FPGGRR15 pKa = 11.84ADD17 pKa = 3.57PTVEE21 pKa = 4.05LVEE24 pKa = 4.27SLARR28 pKa = 11.84YY29 pKa = 8.91ANEE32 pKa = 3.81GFRR35 pKa = 11.84VHH37 pKa = 5.8TWTPGILARR46 pKa = 11.84SFIPVKK52 pKa = 10.57RR53 pKa = 11.84FIDD56 pKa = 3.64AVRR59 pKa = 11.84VLNNEE64 pKa = 3.6PDD66 pKa = 3.27EE67 pKa = 4.47VVFNRR72 pKa = 11.84LLALGVRR79 pKa = 11.84DD80 pKa = 4.1GWWEE84 pKa = 3.97TTNTCTVSQFYY95 pKa = 10.61KK96 pKa = 9.81FCNWLKK102 pKa = 10.34SAEE105 pKa = 4.09GTTRR109 pKa = 11.84LNEE112 pKa = 3.65IRR114 pKa = 11.84KK115 pKa = 9.16ARR117 pKa = 11.84HH118 pKa = 5.69LEE120 pKa = 4.02KK121 pKa = 10.36KK122 pKa = 9.42AQPGQTLEE130 pKa = 4.18DD131 pKa = 3.66VSLVAALDD139 pKa = 3.71EE140 pKa = 4.34QVAEE144 pKa = 4.15FQQRR148 pKa = 11.84KK149 pKa = 9.34KK150 pKa = 7.62EE151 pKa = 4.04TRR153 pKa = 11.84LEE155 pKa = 4.16FEE157 pKa = 4.73SKK159 pKa = 10.8LIDD162 pKa = 3.78LRR164 pKa = 11.84RR165 pKa = 11.84QIALVQQEE173 pKa = 4.2MSATMKK179 pKa = 10.88AHH181 pKa = 7.26DD182 pKa = 5.01ADD184 pKa = 4.94FSPASIYY191 pKa = 11.03EE192 pKa = 4.01PMDD195 pKa = 4.03DD196 pKa = 4.82LEE198 pKa = 5.15FGEE201 pKa = 5.14ACWNLYY207 pKa = 9.8RR208 pKa = 11.84AEE210 pKa = 4.2CARR213 pKa = 11.84LNQDD217 pKa = 4.0EE218 pKa = 4.98APLDD222 pKa = 4.4DD223 pKa = 5.18GLLEE227 pKa = 4.38AVKK230 pKa = 9.06LTHH233 pKa = 6.73GNAALAMHH241 pKa = 7.49KK242 pKa = 10.95ANFLRR247 pKa = 11.84VGFNRR252 pKa = 11.84NNLKK256 pKa = 10.05HH257 pKa = 6.11WIEE260 pKa = 4.01EE261 pKa = 4.41KK262 pKa = 10.41ILEE265 pKa = 4.7LDD267 pKa = 3.63TVGEE271 pKa = 4.13ARR273 pKa = 11.84RR274 pKa = 11.84ATTFRR279 pKa = 11.84SYY281 pKa = 10.48MATTGGALADD291 pKa = 3.78EE292 pKa = 4.95VRR294 pKa = 11.84AEE296 pKa = 4.12TQTRR300 pKa = 11.84VAAAGGTRR308 pKa = 11.84RR309 pKa = 11.84TLRR312 pKa = 11.84RR313 pKa = 11.84ARR315 pKa = 11.84DD316 pKa = 3.41QVDD319 pKa = 3.59VEE321 pKa = 4.36NVQPSRR327 pKa = 11.84LRR329 pKa = 11.84PRR331 pKa = 11.84VTTRR335 pKa = 11.84DD336 pKa = 3.35TGVEE340 pKa = 3.88TAGALVIRR348 pKa = 11.84DD349 pKa = 3.87RR350 pKa = 11.84ASSSHH355 pKa = 5.53TAPPLAEE362 pKa = 4.53PEE364 pKa = 4.31AVHH367 pKa = 6.53PPPAAPGPGGEE378 pKa = 4.53DD379 pKa = 3.49PPLEE383 pKa = 4.19EE384 pKa = 4.71
MM1 pKa = 7.6SGPSQRR7 pKa = 11.84VVHH10 pKa = 6.16FPGGRR15 pKa = 11.84ADD17 pKa = 3.57PTVEE21 pKa = 4.05LVEE24 pKa = 4.27SLARR28 pKa = 11.84YY29 pKa = 8.91ANEE32 pKa = 3.81GFRR35 pKa = 11.84VHH37 pKa = 5.8TWTPGILARR46 pKa = 11.84SFIPVKK52 pKa = 10.57RR53 pKa = 11.84FIDD56 pKa = 3.64AVRR59 pKa = 11.84VLNNEE64 pKa = 3.6PDD66 pKa = 3.27EE67 pKa = 4.47VVFNRR72 pKa = 11.84LLALGVRR79 pKa = 11.84DD80 pKa = 4.1GWWEE84 pKa = 3.97TTNTCTVSQFYY95 pKa = 10.61KK96 pKa = 9.81FCNWLKK102 pKa = 10.34SAEE105 pKa = 4.09GTTRR109 pKa = 11.84LNEE112 pKa = 3.65IRR114 pKa = 11.84KK115 pKa = 9.16ARR117 pKa = 11.84HH118 pKa = 5.69LEE120 pKa = 4.02KK121 pKa = 10.36KK122 pKa = 9.42AQPGQTLEE130 pKa = 4.18DD131 pKa = 3.66VSLVAALDD139 pKa = 3.71EE140 pKa = 4.34QVAEE144 pKa = 4.15FQQRR148 pKa = 11.84KK149 pKa = 9.34KK150 pKa = 7.62EE151 pKa = 4.04TRR153 pKa = 11.84LEE155 pKa = 4.16FEE157 pKa = 4.73SKK159 pKa = 10.8LIDD162 pKa = 3.78LRR164 pKa = 11.84RR165 pKa = 11.84QIALVQQEE173 pKa = 4.2MSATMKK179 pKa = 10.88AHH181 pKa = 7.26DD182 pKa = 5.01ADD184 pKa = 4.94FSPASIYY191 pKa = 11.03EE192 pKa = 4.01PMDD195 pKa = 4.03DD196 pKa = 4.82LEE198 pKa = 5.15FGEE201 pKa = 5.14ACWNLYY207 pKa = 9.8RR208 pKa = 11.84AEE210 pKa = 4.2CARR213 pKa = 11.84LNQDD217 pKa = 4.0EE218 pKa = 4.98APLDD222 pKa = 4.4DD223 pKa = 5.18GLLEE227 pKa = 4.38AVKK230 pKa = 9.06LTHH233 pKa = 6.73GNAALAMHH241 pKa = 7.49KK242 pKa = 10.95ANFLRR247 pKa = 11.84VGFNRR252 pKa = 11.84NNLKK256 pKa = 10.05HH257 pKa = 6.11WIEE260 pKa = 4.01EE261 pKa = 4.41KK262 pKa = 10.41ILEE265 pKa = 4.7LDD267 pKa = 3.63TVGEE271 pKa = 4.13ARR273 pKa = 11.84RR274 pKa = 11.84ATTFRR279 pKa = 11.84SYY281 pKa = 10.48MATTGGALADD291 pKa = 3.78EE292 pKa = 4.95VRR294 pKa = 11.84AEE296 pKa = 4.12TQTRR300 pKa = 11.84VAAAGGTRR308 pKa = 11.84RR309 pKa = 11.84TLRR312 pKa = 11.84RR313 pKa = 11.84ARR315 pKa = 11.84DD316 pKa = 3.41QVDD319 pKa = 3.59VEE321 pKa = 4.36NVQPSRR327 pKa = 11.84LRR329 pKa = 11.84PRR331 pKa = 11.84VTTRR335 pKa = 11.84DD336 pKa = 3.35TGVEE340 pKa = 3.88TAGALVIRR348 pKa = 11.84DD349 pKa = 3.87RR350 pKa = 11.84ASSSHH355 pKa = 5.53TAPPLAEE362 pKa = 4.53PEE364 pKa = 4.31AVHH367 pKa = 6.53PPPAAPGPGGEE378 pKa = 4.53DD379 pKa = 3.49PPLEE383 pKa = 4.19EE384 pKa = 4.71
Molecular weight: 42.87 kDa
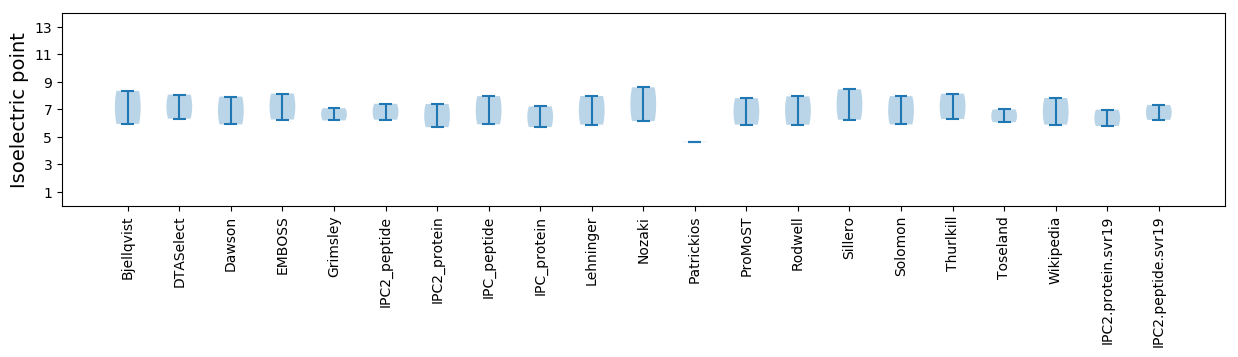
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A3G9M863|A0A3G9M863_9VIRU ORF1+2p OS=Phalaenopsis equestris amalgavirus 1 OX=2069327 GN=ORF1+ORF2 PE=4 SV=1
MM1 pKa = 7.6SGPSQRR7 pKa = 11.84VVHH10 pKa = 6.16FPGGRR15 pKa = 11.84ADD17 pKa = 3.57PTVEE21 pKa = 4.05LVEE24 pKa = 4.27SLARR28 pKa = 11.84YY29 pKa = 8.91ANEE32 pKa = 3.81GFRR35 pKa = 11.84VHH37 pKa = 5.8TWTPGILARR46 pKa = 11.84SFIPVKK52 pKa = 10.57RR53 pKa = 11.84FIDD56 pKa = 3.64AVRR59 pKa = 11.84VLNNEE64 pKa = 3.6PDD66 pKa = 3.27EE67 pKa = 4.47VVFNRR72 pKa = 11.84LLALGVRR79 pKa = 11.84DD80 pKa = 4.1GWWEE84 pKa = 3.97TTNTCTVSQFYY95 pKa = 10.61KK96 pKa = 9.81FCNWLKK102 pKa = 10.34SAEE105 pKa = 4.09GTTRR109 pKa = 11.84LNEE112 pKa = 3.65IRR114 pKa = 11.84KK115 pKa = 9.16ARR117 pKa = 11.84HH118 pKa = 5.69LEE120 pKa = 4.02KK121 pKa = 10.36KK122 pKa = 9.42AQPGQTLEE130 pKa = 4.18DD131 pKa = 3.66VSLVAALDD139 pKa = 3.71EE140 pKa = 4.34QVAEE144 pKa = 4.15FQQRR148 pKa = 11.84KK149 pKa = 9.34KK150 pKa = 7.62EE151 pKa = 4.04TRR153 pKa = 11.84LEE155 pKa = 4.16FEE157 pKa = 4.73SKK159 pKa = 10.8LIDD162 pKa = 3.78LRR164 pKa = 11.84RR165 pKa = 11.84QIALVQQEE173 pKa = 4.2MSATMKK179 pKa = 10.88AHH181 pKa = 7.26DD182 pKa = 5.01ADD184 pKa = 4.94FSPASIYY191 pKa = 11.03EE192 pKa = 4.01PMDD195 pKa = 4.03DD196 pKa = 4.82LEE198 pKa = 5.15FGEE201 pKa = 5.14ACWNLYY207 pKa = 9.8RR208 pKa = 11.84AEE210 pKa = 4.2CARR213 pKa = 11.84LNQDD217 pKa = 4.0EE218 pKa = 4.98APLDD222 pKa = 4.4DD223 pKa = 5.18GLLEE227 pKa = 4.38AVKK230 pKa = 9.06LTHH233 pKa = 6.73GNAALAMHH241 pKa = 7.49KK242 pKa = 10.95ANFLRR247 pKa = 11.84VGFNRR252 pKa = 11.84NNLKK256 pKa = 10.05HH257 pKa = 6.11WIEE260 pKa = 4.01EE261 pKa = 4.41KK262 pKa = 10.41ILEE265 pKa = 4.7LDD267 pKa = 3.63TVGEE271 pKa = 4.13ARR273 pKa = 11.84RR274 pKa = 11.84ATTFVPTWQQRR285 pKa = 11.84VEE287 pKa = 3.6RR288 pKa = 11.84WLMKK292 pKa = 10.54FEE294 pKa = 4.6LKK296 pKa = 10.64LRR298 pKa = 11.84LEE300 pKa = 4.48LLQQVVLGVPCAEE313 pKa = 4.46PGTRR317 pKa = 11.84LTLRR321 pKa = 11.84MSNLLDD327 pKa = 3.88FGLVLRR333 pKa = 11.84RR334 pKa = 11.84EE335 pKa = 4.36ILGSKK340 pKa = 9.69QPAPLSSEE348 pKa = 3.66IEE350 pKa = 4.14RR351 pKa = 11.84AVLIPPLLWQNQRR364 pKa = 11.84LSILRR369 pKa = 11.84PLRR372 pKa = 11.84PDD374 pKa = 3.06LGGRR378 pKa = 11.84IPHH381 pKa = 5.7SRR383 pKa = 11.84SRR385 pKa = 11.84YY386 pKa = 5.37EE387 pKa = 3.63AKK389 pKa = 9.73VRR391 pKa = 11.84KK392 pKa = 9.73VIGGGEE398 pKa = 3.96MRR400 pKa = 11.84DD401 pKa = 3.0WRR403 pKa = 11.84AANAMYY409 pKa = 10.46RR410 pKa = 11.84GGGSFSDD417 pKa = 3.71ALKK420 pKa = 10.95LLIDD424 pKa = 4.15ARR426 pKa = 11.84EE427 pKa = 4.17DD428 pKa = 3.36APGAILSEE436 pKa = 4.03KK437 pKa = 9.65WKK439 pKa = 10.02VDD441 pKa = 3.18SARR444 pKa = 11.84RR445 pKa = 11.84YY446 pKa = 10.21LLLPCGLPVPRR457 pKa = 11.84GPEE460 pKa = 3.57ATKK463 pKa = 10.21MKK465 pKa = 10.54NFNDD469 pKa = 3.98DD470 pKa = 3.2ATAGPALRR478 pKa = 11.84AFGILRR484 pKa = 11.84KK485 pKa = 9.96SGLKK489 pKa = 9.53TSLEE493 pKa = 3.93EE494 pKa = 4.43FAWNCLDD501 pKa = 3.2AFARR505 pKa = 11.84GGAAEE510 pKa = 4.4DD511 pKa = 3.97CLPFVAARR519 pKa = 11.84VGYY522 pKa = 7.78RR523 pKa = 11.84TKK525 pKa = 11.17LLTLSDD531 pKa = 3.67AWSKK535 pKa = 11.14INSCKK540 pKa = 10.1PLGRR544 pKa = 11.84CVMMLDD550 pKa = 3.2AHH552 pKa = 5.87EE553 pKa = 4.36QAFSSPLYY561 pKa = 10.41NVLSNLTHH569 pKa = 7.2LSRR572 pKa = 11.84FQRR575 pKa = 11.84NSGFCNSIVRR585 pKa = 11.84ASSDD589 pKa = 2.63WAMLWKK595 pKa = 10.4DD596 pKa = 3.41VSAASCVVEE605 pKa = 4.82LDD607 pKa = 2.93WKK609 pKa = 10.89KK610 pKa = 10.72FDD612 pKa = 4.24RR613 pKa = 11.84EE614 pKa = 4.03RR615 pKa = 11.84PADD618 pKa = 4.95DD619 pKa = 3.34ISFMIDD625 pKa = 3.13VIISCFEE632 pKa = 4.0AKK634 pKa = 10.07DD635 pKa = 3.78DD636 pKa = 3.99YY637 pKa = 11.47EE638 pKa = 4.11EE639 pKa = 4.6RR640 pKa = 11.84LLLGYY645 pKa = 10.55RR646 pKa = 11.84IMLNRR651 pKa = 11.84ALIEE655 pKa = 4.2RR656 pKa = 11.84CFVTDD661 pKa = 3.67DD662 pKa = 3.76GGVFHH667 pKa = 7.72IDD669 pKa = 2.91GMVPSGSLWTGWLDD683 pKa = 3.31TALNILYY690 pKa = 9.99IGAALRR696 pKa = 11.84HH697 pKa = 5.45VLPDD701 pKa = 3.57TSQAVAKK708 pKa = 10.53CAGDD712 pKa = 4.04DD713 pKa = 3.86NLTLFYY719 pKa = 10.67TDD721 pKa = 5.42LPDD724 pKa = 5.2ASLLNLKK731 pKa = 10.19KK732 pKa = 10.71YY733 pKa = 9.83LNEE736 pKa = 3.57WFRR739 pKa = 11.84AGIEE743 pKa = 4.09DD744 pKa = 3.91EE745 pKa = 4.96DD746 pKa = 5.19FIIHH750 pKa = 6.73RR751 pKa = 11.84PPYY754 pKa = 10.23HH755 pKa = 5.77ITRR758 pKa = 11.84FQATFPPGTDD768 pKa = 3.12LSKK771 pKa = 10.61GTSHH775 pKa = 7.51LLDD778 pKa = 3.45SAKK781 pKa = 9.85WIQIHH786 pKa = 6.74GIMNIDD792 pKa = 3.4EE793 pKa = 4.53AAGLSHH799 pKa = 6.66RR800 pKa = 11.84WKK802 pKa = 10.74YY803 pKa = 11.22SFAGKK808 pKa = 9.41PKK810 pKa = 10.27FLSCYY815 pKa = 8.77WEE817 pKa = 4.29EE818 pKa = 3.98NGNPIRR824 pKa = 11.84PAHH827 pKa = 6.0VNLEE831 pKa = 4.01KK832 pKa = 10.97LLWPEE837 pKa = 5.32GIHH840 pKa = 6.05KK841 pKa = 9.4TIDD844 pKa = 3.47DD845 pKa = 3.85YY846 pKa = 11.81LAAVISMVVDD856 pKa = 3.37NPFNHH861 pKa = 6.68HH862 pKa = 5.94NVNHH866 pKa = 5.55MMHH869 pKa = 7.34RR870 pKa = 11.84FCIIQQVKK878 pKa = 8.77RR879 pKa = 11.84LSVAGIRR886 pKa = 11.84DD887 pKa = 3.68DD888 pKa = 5.43HH889 pKa = 7.41ILSLAHH895 pKa = 6.37IRR897 pKa = 11.84GKK899 pKa = 10.05EE900 pKa = 3.95GEE902 pKa = 4.4MVPFPMVAEE911 pKa = 4.29WRR913 pKa = 11.84RR914 pKa = 11.84TQGWVDD920 pKa = 3.95MEE922 pKa = 4.57SMPYY926 pKa = 8.98LKK928 pKa = 10.29KK929 pKa = 10.49YY930 pKa = 10.58ISDD933 pKa = 3.66FRR935 pKa = 11.84HH936 pKa = 5.63FAAGVMSLYY945 pKa = 10.64ARR947 pKa = 11.84QPGGGIDD954 pKa = 3.0AWRR957 pKa = 11.84FMDD960 pKa = 4.28MIRR963 pKa = 11.84GNVLLGGEE971 pKa = 4.05QFGNDD976 pKa = 2.81FRR978 pKa = 11.84DD979 pKa = 3.6WIKK982 pKa = 10.53FLQNNPLTSYY992 pKa = 9.4LKK994 pKa = 9.71PMRR997 pKa = 11.84RR998 pKa = 11.84FRR1000 pKa = 11.84PQKK1003 pKa = 9.56AARR1006 pKa = 11.84EE1007 pKa = 4.17STEE1010 pKa = 4.92DD1011 pKa = 3.37IISKK1015 pKa = 10.0FKK1017 pKa = 11.08SFSACCGIGDD1027 pKa = 4.01GRR1029 pKa = 11.84TPFASTEE1036 pKa = 4.4SYY1038 pKa = 10.99GSWIADD1044 pKa = 3.4LVPRR1048 pKa = 11.84MQHH1051 pKa = 5.1GKK1053 pKa = 10.62APLATMM1059 pKa = 5.09
MM1 pKa = 7.6SGPSQRR7 pKa = 11.84VVHH10 pKa = 6.16FPGGRR15 pKa = 11.84ADD17 pKa = 3.57PTVEE21 pKa = 4.05LVEE24 pKa = 4.27SLARR28 pKa = 11.84YY29 pKa = 8.91ANEE32 pKa = 3.81GFRR35 pKa = 11.84VHH37 pKa = 5.8TWTPGILARR46 pKa = 11.84SFIPVKK52 pKa = 10.57RR53 pKa = 11.84FIDD56 pKa = 3.64AVRR59 pKa = 11.84VLNNEE64 pKa = 3.6PDD66 pKa = 3.27EE67 pKa = 4.47VVFNRR72 pKa = 11.84LLALGVRR79 pKa = 11.84DD80 pKa = 4.1GWWEE84 pKa = 3.97TTNTCTVSQFYY95 pKa = 10.61KK96 pKa = 9.81FCNWLKK102 pKa = 10.34SAEE105 pKa = 4.09GTTRR109 pKa = 11.84LNEE112 pKa = 3.65IRR114 pKa = 11.84KK115 pKa = 9.16ARR117 pKa = 11.84HH118 pKa = 5.69LEE120 pKa = 4.02KK121 pKa = 10.36KK122 pKa = 9.42AQPGQTLEE130 pKa = 4.18DD131 pKa = 3.66VSLVAALDD139 pKa = 3.71EE140 pKa = 4.34QVAEE144 pKa = 4.15FQQRR148 pKa = 11.84KK149 pKa = 9.34KK150 pKa = 7.62EE151 pKa = 4.04TRR153 pKa = 11.84LEE155 pKa = 4.16FEE157 pKa = 4.73SKK159 pKa = 10.8LIDD162 pKa = 3.78LRR164 pKa = 11.84RR165 pKa = 11.84QIALVQQEE173 pKa = 4.2MSATMKK179 pKa = 10.88AHH181 pKa = 7.26DD182 pKa = 5.01ADD184 pKa = 4.94FSPASIYY191 pKa = 11.03EE192 pKa = 4.01PMDD195 pKa = 4.03DD196 pKa = 4.82LEE198 pKa = 5.15FGEE201 pKa = 5.14ACWNLYY207 pKa = 9.8RR208 pKa = 11.84AEE210 pKa = 4.2CARR213 pKa = 11.84LNQDD217 pKa = 4.0EE218 pKa = 4.98APLDD222 pKa = 4.4DD223 pKa = 5.18GLLEE227 pKa = 4.38AVKK230 pKa = 9.06LTHH233 pKa = 6.73GNAALAMHH241 pKa = 7.49KK242 pKa = 10.95ANFLRR247 pKa = 11.84VGFNRR252 pKa = 11.84NNLKK256 pKa = 10.05HH257 pKa = 6.11WIEE260 pKa = 4.01EE261 pKa = 4.41KK262 pKa = 10.41ILEE265 pKa = 4.7LDD267 pKa = 3.63TVGEE271 pKa = 4.13ARR273 pKa = 11.84RR274 pKa = 11.84ATTFVPTWQQRR285 pKa = 11.84VEE287 pKa = 3.6RR288 pKa = 11.84WLMKK292 pKa = 10.54FEE294 pKa = 4.6LKK296 pKa = 10.64LRR298 pKa = 11.84LEE300 pKa = 4.48LLQQVVLGVPCAEE313 pKa = 4.46PGTRR317 pKa = 11.84LTLRR321 pKa = 11.84MSNLLDD327 pKa = 3.88FGLVLRR333 pKa = 11.84RR334 pKa = 11.84EE335 pKa = 4.36ILGSKK340 pKa = 9.69QPAPLSSEE348 pKa = 3.66IEE350 pKa = 4.14RR351 pKa = 11.84AVLIPPLLWQNQRR364 pKa = 11.84LSILRR369 pKa = 11.84PLRR372 pKa = 11.84PDD374 pKa = 3.06LGGRR378 pKa = 11.84IPHH381 pKa = 5.7SRR383 pKa = 11.84SRR385 pKa = 11.84YY386 pKa = 5.37EE387 pKa = 3.63AKK389 pKa = 9.73VRR391 pKa = 11.84KK392 pKa = 9.73VIGGGEE398 pKa = 3.96MRR400 pKa = 11.84DD401 pKa = 3.0WRR403 pKa = 11.84AANAMYY409 pKa = 10.46RR410 pKa = 11.84GGGSFSDD417 pKa = 3.71ALKK420 pKa = 10.95LLIDD424 pKa = 4.15ARR426 pKa = 11.84EE427 pKa = 4.17DD428 pKa = 3.36APGAILSEE436 pKa = 4.03KK437 pKa = 9.65WKK439 pKa = 10.02VDD441 pKa = 3.18SARR444 pKa = 11.84RR445 pKa = 11.84YY446 pKa = 10.21LLLPCGLPVPRR457 pKa = 11.84GPEE460 pKa = 3.57ATKK463 pKa = 10.21MKK465 pKa = 10.54NFNDD469 pKa = 3.98DD470 pKa = 3.2ATAGPALRR478 pKa = 11.84AFGILRR484 pKa = 11.84KK485 pKa = 9.96SGLKK489 pKa = 9.53TSLEE493 pKa = 3.93EE494 pKa = 4.43FAWNCLDD501 pKa = 3.2AFARR505 pKa = 11.84GGAAEE510 pKa = 4.4DD511 pKa = 3.97CLPFVAARR519 pKa = 11.84VGYY522 pKa = 7.78RR523 pKa = 11.84TKK525 pKa = 11.17LLTLSDD531 pKa = 3.67AWSKK535 pKa = 11.14INSCKK540 pKa = 10.1PLGRR544 pKa = 11.84CVMMLDD550 pKa = 3.2AHH552 pKa = 5.87EE553 pKa = 4.36QAFSSPLYY561 pKa = 10.41NVLSNLTHH569 pKa = 7.2LSRR572 pKa = 11.84FQRR575 pKa = 11.84NSGFCNSIVRR585 pKa = 11.84ASSDD589 pKa = 2.63WAMLWKK595 pKa = 10.4DD596 pKa = 3.41VSAASCVVEE605 pKa = 4.82LDD607 pKa = 2.93WKK609 pKa = 10.89KK610 pKa = 10.72FDD612 pKa = 4.24RR613 pKa = 11.84EE614 pKa = 4.03RR615 pKa = 11.84PADD618 pKa = 4.95DD619 pKa = 3.34ISFMIDD625 pKa = 3.13VIISCFEE632 pKa = 4.0AKK634 pKa = 10.07DD635 pKa = 3.78DD636 pKa = 3.99YY637 pKa = 11.47EE638 pKa = 4.11EE639 pKa = 4.6RR640 pKa = 11.84LLLGYY645 pKa = 10.55RR646 pKa = 11.84IMLNRR651 pKa = 11.84ALIEE655 pKa = 4.2RR656 pKa = 11.84CFVTDD661 pKa = 3.67DD662 pKa = 3.76GGVFHH667 pKa = 7.72IDD669 pKa = 2.91GMVPSGSLWTGWLDD683 pKa = 3.31TALNILYY690 pKa = 9.99IGAALRR696 pKa = 11.84HH697 pKa = 5.45VLPDD701 pKa = 3.57TSQAVAKK708 pKa = 10.53CAGDD712 pKa = 4.04DD713 pKa = 3.86NLTLFYY719 pKa = 10.67TDD721 pKa = 5.42LPDD724 pKa = 5.2ASLLNLKK731 pKa = 10.19KK732 pKa = 10.71YY733 pKa = 9.83LNEE736 pKa = 3.57WFRR739 pKa = 11.84AGIEE743 pKa = 4.09DD744 pKa = 3.91EE745 pKa = 4.96DD746 pKa = 5.19FIIHH750 pKa = 6.73RR751 pKa = 11.84PPYY754 pKa = 10.23HH755 pKa = 5.77ITRR758 pKa = 11.84FQATFPPGTDD768 pKa = 3.12LSKK771 pKa = 10.61GTSHH775 pKa = 7.51LLDD778 pKa = 3.45SAKK781 pKa = 9.85WIQIHH786 pKa = 6.74GIMNIDD792 pKa = 3.4EE793 pKa = 4.53AAGLSHH799 pKa = 6.66RR800 pKa = 11.84WKK802 pKa = 10.74YY803 pKa = 11.22SFAGKK808 pKa = 9.41PKK810 pKa = 10.27FLSCYY815 pKa = 8.77WEE817 pKa = 4.29EE818 pKa = 3.98NGNPIRR824 pKa = 11.84PAHH827 pKa = 6.0VNLEE831 pKa = 4.01KK832 pKa = 10.97LLWPEE837 pKa = 5.32GIHH840 pKa = 6.05KK841 pKa = 9.4TIDD844 pKa = 3.47DD845 pKa = 3.85YY846 pKa = 11.81LAAVISMVVDD856 pKa = 3.37NPFNHH861 pKa = 6.68HH862 pKa = 5.94NVNHH866 pKa = 5.55MMHH869 pKa = 7.34RR870 pKa = 11.84FCIIQQVKK878 pKa = 8.77RR879 pKa = 11.84LSVAGIRR886 pKa = 11.84DD887 pKa = 3.68DD888 pKa = 5.43HH889 pKa = 7.41ILSLAHH895 pKa = 6.37IRR897 pKa = 11.84GKK899 pKa = 10.05EE900 pKa = 3.95GEE902 pKa = 4.4MVPFPMVAEE911 pKa = 4.29WRR913 pKa = 11.84RR914 pKa = 11.84TQGWVDD920 pKa = 3.95MEE922 pKa = 4.57SMPYY926 pKa = 8.98LKK928 pKa = 10.29KK929 pKa = 10.49YY930 pKa = 10.58ISDD933 pKa = 3.66FRR935 pKa = 11.84HH936 pKa = 5.63FAAGVMSLYY945 pKa = 10.64ARR947 pKa = 11.84QPGGGIDD954 pKa = 3.0AWRR957 pKa = 11.84FMDD960 pKa = 4.28MIRR963 pKa = 11.84GNVLLGGEE971 pKa = 4.05QFGNDD976 pKa = 2.81FRR978 pKa = 11.84DD979 pKa = 3.6WIKK982 pKa = 10.53FLQNNPLTSYY992 pKa = 9.4LKK994 pKa = 9.71PMRR997 pKa = 11.84RR998 pKa = 11.84FRR1000 pKa = 11.84PQKK1003 pKa = 9.56AARR1006 pKa = 11.84EE1007 pKa = 4.17STEE1010 pKa = 4.92DD1011 pKa = 3.37IISKK1015 pKa = 10.0FKK1017 pKa = 11.08SFSACCGIGDD1027 pKa = 4.01GRR1029 pKa = 11.84TPFASTEE1036 pKa = 4.4SYY1038 pKa = 10.99GSWIADD1044 pKa = 3.4LVPRR1048 pKa = 11.84MQHH1051 pKa = 5.1GKK1053 pKa = 10.62APLATMM1059 pKa = 5.09
Molecular weight: 120.14 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1443 |
384 |
1059 |
721.5 |
81.51 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
9.425 ± 0.911 | 1.594 ± 0.248 |
5.96 ± 0.22 | 6.861 ± 1.01 |
4.435 ± 0.354 | 6.445 ± 0.204 |
2.495 ± 0.068 | 4.435 ± 0.821 |
4.643 ± 0.447 | 10.395 ± 0.574 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.495 ± 0.418 | 3.95 ± 0.02 |
5.128 ± 0.269 | 3.119 ± 0.236 |
8.177 ± 0.537 | 5.475 ± 0.586 |
4.782 ± 1.008 | 5.891 ± 0.628 |
2.356 ± 0.356 | 1.94 ± 0.286 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
