
Acholeplasma brassicae
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Tenericutes; Mollicutes; Acholeplasmatales; Acholeplasmataceae; Acholeplasma
Average proteome isoelectric point is 6.55
Get precalculated fractions of proteins
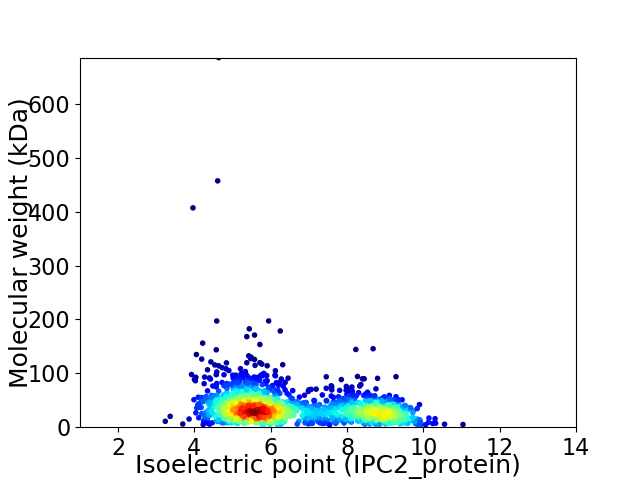
Virtual 2D-PAGE plot for 1671 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|U4KQ71|U4KQ71_9MOLU Pyrroline-5-carboxylate reductase OS=Acholeplasma brassicae OX=61635 GN=proC PE=3 SV=1
MM1 pKa = 7.6NEE3 pKa = 3.95EE4 pKa = 4.96AILTYY9 pKa = 10.18TVGDD13 pKa = 3.93PFSDD17 pKa = 3.89IIASTWCEE25 pKa = 3.9GIDD28 pKa = 4.05NVDD31 pKa = 3.38EE32 pKa = 4.35TVDD35 pKa = 4.72CEE37 pKa = 4.24FLSHH41 pKa = 6.93GIVNTSIAGTYY52 pKa = 9.38ILIYY56 pKa = 10.22QATDD60 pKa = 2.98NAGNTAEE67 pKa = 4.52LRR69 pKa = 11.84LTVTVSDD76 pKa = 4.12VVEE79 pKa = 4.48SNPDD83 pKa = 3.04VLAYY87 pKa = 10.25YY88 pKa = 10.58SSAEE92 pKa = 3.99GLSGNTLFLEE102 pKa = 4.43LRR104 pKa = 11.84SIIQADD110 pKa = 4.23MIKK113 pKa = 10.53VSYY116 pKa = 10.97SDD118 pKa = 3.0ARR120 pKa = 11.84YY121 pKa = 9.83ILDD124 pKa = 5.02EE125 pKa = 5.24ADD127 pKa = 3.23QDD129 pKa = 4.27PNNSNNVLTIYY140 pKa = 10.45DD141 pKa = 3.83RR142 pKa = 11.84QSVLGAWDD150 pKa = 3.28GTTYY154 pKa = 10.14TRR156 pKa = 11.84EE157 pKa = 4.24HH158 pKa = 5.59VWPNSRR164 pKa = 11.84LGVSRR169 pKa = 11.84VSNSTKK175 pKa = 10.77NIGTDD180 pKa = 3.18LHH182 pKa = 6.28NLRR185 pKa = 11.84ATIQSTNSSRR195 pKa = 11.84SNKK198 pKa = 9.67YY199 pKa = 10.44FDD201 pKa = 3.51FTTTNDD207 pKa = 2.91AYY209 pKa = 11.53YY210 pKa = 10.08PGEE213 pKa = 4.29DD214 pKa = 4.17DD215 pKa = 4.08KK216 pKa = 12.16GDD218 pKa = 3.36VARR221 pKa = 11.84ILFYY225 pKa = 10.54MVVMYY230 pKa = 10.04PNLDD234 pKa = 3.16IVNVITSAMDD244 pKa = 3.11EE245 pKa = 4.14ATYY248 pKa = 10.89KK249 pKa = 10.91EE250 pKa = 4.82DD251 pKa = 3.3GTYY254 pKa = 7.79MAKK257 pKa = 9.67MSVLLQWHH265 pKa = 6.66IEE267 pKa = 4.09DD268 pKa = 4.43PVDD271 pKa = 3.9DD272 pKa = 4.36FEE274 pKa = 5.95RR275 pKa = 11.84NRR277 pKa = 11.84NEE279 pKa = 4.49VIYY282 pKa = 10.49NYY284 pKa = 10.31QNNRR288 pKa = 11.84NPFIDD293 pKa = 3.72NPDD296 pKa = 3.4YY297 pKa = 11.32VALLWGNNPSTVSNSSQFINN317 pKa = 3.72
MM1 pKa = 7.6NEE3 pKa = 3.95EE4 pKa = 4.96AILTYY9 pKa = 10.18TVGDD13 pKa = 3.93PFSDD17 pKa = 3.89IIASTWCEE25 pKa = 3.9GIDD28 pKa = 4.05NVDD31 pKa = 3.38EE32 pKa = 4.35TVDD35 pKa = 4.72CEE37 pKa = 4.24FLSHH41 pKa = 6.93GIVNTSIAGTYY52 pKa = 9.38ILIYY56 pKa = 10.22QATDD60 pKa = 2.98NAGNTAEE67 pKa = 4.52LRR69 pKa = 11.84LTVTVSDD76 pKa = 4.12VVEE79 pKa = 4.48SNPDD83 pKa = 3.04VLAYY87 pKa = 10.25YY88 pKa = 10.58SSAEE92 pKa = 3.99GLSGNTLFLEE102 pKa = 4.43LRR104 pKa = 11.84SIIQADD110 pKa = 4.23MIKK113 pKa = 10.53VSYY116 pKa = 10.97SDD118 pKa = 3.0ARR120 pKa = 11.84YY121 pKa = 9.83ILDD124 pKa = 5.02EE125 pKa = 5.24ADD127 pKa = 3.23QDD129 pKa = 4.27PNNSNNVLTIYY140 pKa = 10.45DD141 pKa = 3.83RR142 pKa = 11.84QSVLGAWDD150 pKa = 3.28GTTYY154 pKa = 10.14TRR156 pKa = 11.84EE157 pKa = 4.24HH158 pKa = 5.59VWPNSRR164 pKa = 11.84LGVSRR169 pKa = 11.84VSNSTKK175 pKa = 10.77NIGTDD180 pKa = 3.18LHH182 pKa = 6.28NLRR185 pKa = 11.84ATIQSTNSSRR195 pKa = 11.84SNKK198 pKa = 9.67YY199 pKa = 10.44FDD201 pKa = 3.51FTTTNDD207 pKa = 2.91AYY209 pKa = 11.53YY210 pKa = 10.08PGEE213 pKa = 4.29DD214 pKa = 4.17DD215 pKa = 4.08KK216 pKa = 12.16GDD218 pKa = 3.36VARR221 pKa = 11.84ILFYY225 pKa = 10.54MVVMYY230 pKa = 10.04PNLDD234 pKa = 3.16IVNVITSAMDD244 pKa = 3.11EE245 pKa = 4.14ATYY248 pKa = 10.89KK249 pKa = 10.91EE250 pKa = 4.82DD251 pKa = 3.3GTYY254 pKa = 7.79MAKK257 pKa = 9.67MSVLLQWHH265 pKa = 6.66IEE267 pKa = 4.09DD268 pKa = 4.43PVDD271 pKa = 3.9DD272 pKa = 4.36FEE274 pKa = 5.95RR275 pKa = 11.84NRR277 pKa = 11.84NEE279 pKa = 4.49VIYY282 pKa = 10.49NYY284 pKa = 10.31QNNRR288 pKa = 11.84NPFIDD293 pKa = 3.72NPDD296 pKa = 3.4YY297 pKa = 11.32VALLWGNNPSTVSNSSQFINN317 pKa = 3.72
Molecular weight: 35.64 kDa
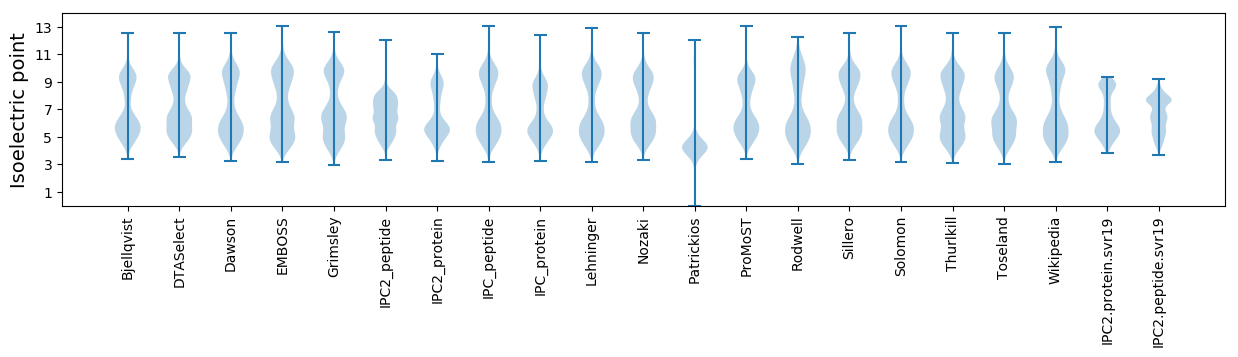
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|U4KQR7|U4KQR7_9MOLU Bifunctional protein GlmU OS=Acholeplasma brassicae OX=61635 GN=glmU PE=3 SV=1
MM1 pKa = 7.35KK2 pKa = 9.43RR3 pKa = 11.84TYY5 pKa = 10.28QPSKK9 pKa = 10.61LKK11 pKa = 10.22RR12 pKa = 11.84QKK14 pKa = 8.58THH16 pKa = 5.13GFRR19 pKa = 11.84ARR21 pKa = 11.84MATVGGRR28 pKa = 11.84AVLARR33 pKa = 11.84RR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 10.4GRR39 pKa = 11.84ASLTVV44 pKa = 3.04
MM1 pKa = 7.35KK2 pKa = 9.43RR3 pKa = 11.84TYY5 pKa = 10.28QPSKK9 pKa = 10.61LKK11 pKa = 10.22RR12 pKa = 11.84QKK14 pKa = 8.58THH16 pKa = 5.13GFRR19 pKa = 11.84ARR21 pKa = 11.84MATVGGRR28 pKa = 11.84AVLARR33 pKa = 11.84RR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 10.4GRR39 pKa = 11.84ASLTVV44 pKa = 3.04
Molecular weight: 5.0 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
559421 |
37 |
6236 |
334.8 |
38.03 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
5.528 ± 0.062 | 0.53 ± 0.016 |
5.749 ± 0.049 | 6.714 ± 0.054 |
4.987 ± 0.052 | 5.886 ± 0.069 |
1.853 ± 0.029 | 8.547 ± 0.067 |
7.744 ± 0.074 | 10.272 ± 0.069 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.528 ± 0.037 | 5.389 ± 0.049 |
2.845 ± 0.027 | 3.455 ± 0.031 |
3.588 ± 0.038 | 6.37 ± 0.057 |
5.664 ± 0.056 | 7.038 ± 0.056 |
0.657 ± 0.019 | 4.658 ± 0.059 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
