
Microbacterium chocolatum
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Actinobacteria;
Average proteome isoelectric point is 6.01
Get precalculated fractions of proteins
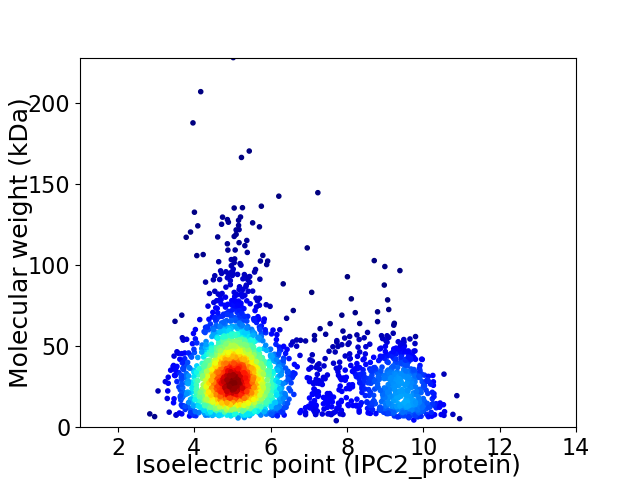
Virtual 2D-PAGE plot for 2721 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
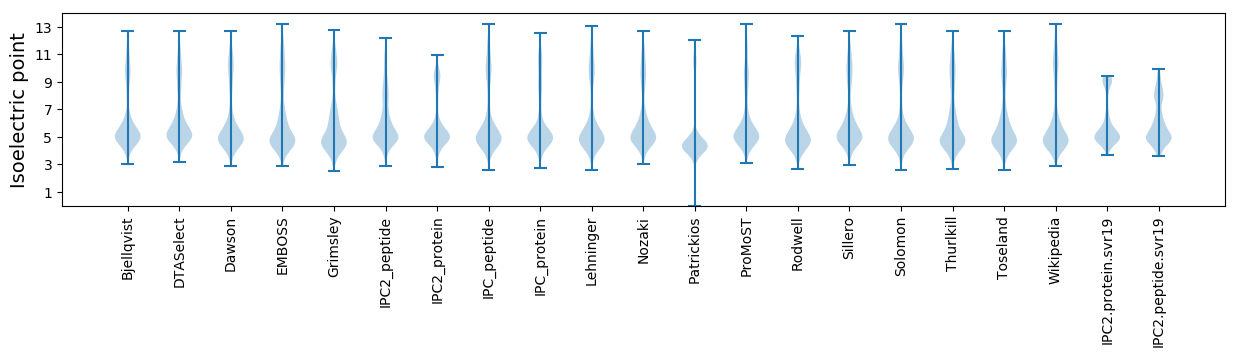
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A0M8MMV9|A0A0M8MMV9_9MICO Abasic site processing protein OS=Microbacterium chocolatum OX=84292 GN=XI38_12245 PE=3 SV=1
MM1 pKa = 7.58LAPAATLALGALVLAGCAGGAPASSSAPGEE31 pKa = 4.09ADD33 pKa = 3.16PQAEE37 pKa = 4.48VIVGSQNEE45 pKa = 4.43PTNLDD50 pKa = 3.56QIFGGSSGVTEE61 pKa = 4.12VFTGNVYY68 pKa = 10.71EE69 pKa = 4.49GLFKK73 pKa = 10.27ITDD76 pKa = 3.77DD77 pKa = 4.04AAVEE81 pKa = 4.06PLLATDD87 pKa = 4.22TEE89 pKa = 4.42VSEE92 pKa = 5.21DD93 pKa = 3.39GLVYY97 pKa = 10.54TFTLQPDD104 pKa = 3.75ATFHH108 pKa = 6.98SGDD111 pKa = 3.78PVDD114 pKa = 5.03AEE116 pKa = 4.05AVKK119 pKa = 10.87YY120 pKa = 10.22SLEE123 pKa = 4.11RR124 pKa = 11.84FVSEE128 pKa = 3.75EE129 pKa = 3.76SIAARR134 pKa = 11.84KK135 pKa = 8.78RR136 pKa = 11.84QLSVIDD142 pKa = 3.85SVEE145 pKa = 4.23VIDD148 pKa = 5.0DD149 pKa = 3.49RR150 pKa = 11.84TVAVTLSQPSISFTYY165 pKa = 9.69NLGYY169 pKa = 10.53VWIVNPAAGDD179 pKa = 3.65LTSAADD185 pKa = 3.78GSGPYY190 pKa = 10.6ALADD194 pKa = 3.61YY195 pKa = 10.76RR196 pKa = 11.84KK197 pKa = 9.24GDD199 pKa = 4.03SIALEE204 pKa = 4.19VNEE207 pKa = 4.98EE208 pKa = 4.27YY209 pKa = 10.02WGEE212 pKa = 3.85APANGGVVYY221 pKa = 10.11QYY223 pKa = 11.22YY224 pKa = 11.08ADD226 pKa = 3.7PTALNNALLTNAVDD240 pKa = 4.68LVTSQSNPDD249 pKa = 3.24SLAEE253 pKa = 3.81FEE255 pKa = 4.35AAGFEE260 pKa = 4.45IIEE263 pKa = 4.47GTSTTKK269 pKa = 10.62EE270 pKa = 3.52LLAFNDD276 pKa = 4.74RR277 pKa = 11.84IAPFDD282 pKa = 4.97DD283 pKa = 3.21IQVRR287 pKa = 11.84KK288 pKa = 9.98AIYY291 pKa = 9.5SAINRR296 pKa = 11.84EE297 pKa = 3.98QLLDD301 pKa = 4.87AIWDD305 pKa = 3.71GRR307 pKa = 11.84GQLIGSMVPPSEE319 pKa = 3.77PWYY322 pKa = 11.0LDD324 pKa = 3.57LADD327 pKa = 4.63NNPYY331 pKa = 10.79DD332 pKa = 3.84PALSEE337 pKa = 4.5EE338 pKa = 4.37LLADD342 pKa = 4.02AGYY345 pKa = 11.47ADD347 pKa = 4.53GFSFTLDD354 pKa = 3.47TPDD357 pKa = 3.65SGVHH361 pKa = 4.14STVAEE366 pKa = 4.24FVKK369 pKa = 10.42TEE371 pKa = 3.95LAKK374 pKa = 11.02VGITVDD380 pKa = 3.24INIITDD386 pKa = 3.59DD387 pKa = 2.98EE388 pKa = 4.96WYY390 pKa = 10.5QKK392 pKa = 11.05VYY394 pKa = 10.79TDD396 pKa = 3.59TDD398 pKa = 3.99FEE400 pKa = 4.38ATLQGHH406 pKa = 5.63VNDD409 pKa = 4.96RR410 pKa = 11.84DD411 pKa = 3.37INFYY415 pKa = 11.17GNPDD419 pKa = 4.21FYY421 pKa = 10.66WGYY424 pKa = 11.37DD425 pKa = 3.45NADD428 pKa = 3.48VQTWLADD435 pKa = 3.46SEE437 pKa = 4.4AASTVEE443 pKa = 4.08EE444 pKa = 3.95QTEE447 pKa = 4.27LVKK450 pKa = 10.75LANQQISDD458 pKa = 3.92DD459 pKa = 3.97AASVWLYY466 pKa = 10.7LNPQLRR472 pKa = 11.84VAAPGIGGVPEE483 pKa = 4.34NGLNSLFYY491 pKa = 11.06VYY493 pKa = 10.11DD494 pKa = 3.47IEE496 pKa = 4.87KK497 pKa = 10.46AASS500 pKa = 3.16
MM1 pKa = 7.58LAPAATLALGALVLAGCAGGAPASSSAPGEE31 pKa = 4.09ADD33 pKa = 3.16PQAEE37 pKa = 4.48VIVGSQNEE45 pKa = 4.43PTNLDD50 pKa = 3.56QIFGGSSGVTEE61 pKa = 4.12VFTGNVYY68 pKa = 10.71EE69 pKa = 4.49GLFKK73 pKa = 10.27ITDD76 pKa = 3.77DD77 pKa = 4.04AAVEE81 pKa = 4.06PLLATDD87 pKa = 4.22TEE89 pKa = 4.42VSEE92 pKa = 5.21DD93 pKa = 3.39GLVYY97 pKa = 10.54TFTLQPDD104 pKa = 3.75ATFHH108 pKa = 6.98SGDD111 pKa = 3.78PVDD114 pKa = 5.03AEE116 pKa = 4.05AVKK119 pKa = 10.87YY120 pKa = 10.22SLEE123 pKa = 4.11RR124 pKa = 11.84FVSEE128 pKa = 3.75EE129 pKa = 3.76SIAARR134 pKa = 11.84KK135 pKa = 8.78RR136 pKa = 11.84QLSVIDD142 pKa = 3.85SVEE145 pKa = 4.23VIDD148 pKa = 5.0DD149 pKa = 3.49RR150 pKa = 11.84TVAVTLSQPSISFTYY165 pKa = 9.69NLGYY169 pKa = 10.53VWIVNPAAGDD179 pKa = 3.65LTSAADD185 pKa = 3.78GSGPYY190 pKa = 10.6ALADD194 pKa = 3.61YY195 pKa = 10.76RR196 pKa = 11.84KK197 pKa = 9.24GDD199 pKa = 4.03SIALEE204 pKa = 4.19VNEE207 pKa = 4.98EE208 pKa = 4.27YY209 pKa = 10.02WGEE212 pKa = 3.85APANGGVVYY221 pKa = 10.11QYY223 pKa = 11.22YY224 pKa = 11.08ADD226 pKa = 3.7PTALNNALLTNAVDD240 pKa = 4.68LVTSQSNPDD249 pKa = 3.24SLAEE253 pKa = 3.81FEE255 pKa = 4.35AAGFEE260 pKa = 4.45IIEE263 pKa = 4.47GTSTTKK269 pKa = 10.62EE270 pKa = 3.52LLAFNDD276 pKa = 4.74RR277 pKa = 11.84IAPFDD282 pKa = 4.97DD283 pKa = 3.21IQVRR287 pKa = 11.84KK288 pKa = 9.98AIYY291 pKa = 9.5SAINRR296 pKa = 11.84EE297 pKa = 3.98QLLDD301 pKa = 4.87AIWDD305 pKa = 3.71GRR307 pKa = 11.84GQLIGSMVPPSEE319 pKa = 3.77PWYY322 pKa = 11.0LDD324 pKa = 3.57LADD327 pKa = 4.63NNPYY331 pKa = 10.79DD332 pKa = 3.84PALSEE337 pKa = 4.5EE338 pKa = 4.37LLADD342 pKa = 4.02AGYY345 pKa = 11.47ADD347 pKa = 4.53GFSFTLDD354 pKa = 3.47TPDD357 pKa = 3.65SGVHH361 pKa = 4.14STVAEE366 pKa = 4.24FVKK369 pKa = 10.42TEE371 pKa = 3.95LAKK374 pKa = 11.02VGITVDD380 pKa = 3.24INIITDD386 pKa = 3.59DD387 pKa = 2.98EE388 pKa = 4.96WYY390 pKa = 10.5QKK392 pKa = 11.05VYY394 pKa = 10.79TDD396 pKa = 3.59TDD398 pKa = 3.99FEE400 pKa = 4.38ATLQGHH406 pKa = 5.63VNDD409 pKa = 4.96RR410 pKa = 11.84DD411 pKa = 3.37INFYY415 pKa = 11.17GNPDD419 pKa = 4.21FYY421 pKa = 10.66WGYY424 pKa = 11.37DD425 pKa = 3.45NADD428 pKa = 3.48VQTWLADD435 pKa = 3.46SEE437 pKa = 4.4AASTVEE443 pKa = 4.08EE444 pKa = 3.95QTEE447 pKa = 4.27LVKK450 pKa = 10.75LANQQISDD458 pKa = 3.92DD459 pKa = 3.97AASVWLYY466 pKa = 10.7LNPQLRR472 pKa = 11.84VAAPGIGGVPEE483 pKa = 4.34NGLNSLFYY491 pKa = 11.06VYY493 pKa = 10.11DD494 pKa = 3.47IEE496 pKa = 4.87KK497 pKa = 10.46AASS500 pKa = 3.16
Molecular weight: 53.7 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A0M8MQM2|A0A0M8MQM2_9MICO LacI family transcriptional regulator OS=Microbacterium chocolatum OX=84292 GN=XI38_00950 PE=4 SV=1
MM1 pKa = 7.69SKK3 pKa = 9.0RR4 pKa = 11.84TFQPNNRR11 pKa = 11.84RR12 pKa = 11.84RR13 pKa = 11.84AKK15 pKa = 10.04KK16 pKa = 8.86HH17 pKa = 4.36GFRR20 pKa = 11.84ARR22 pKa = 11.84MRR24 pKa = 11.84TRR26 pKa = 11.84AGRR29 pKa = 11.84SILAARR35 pKa = 11.84RR36 pKa = 11.84SKK38 pKa = 10.97GRR40 pKa = 11.84TEE42 pKa = 4.38LSAA45 pKa = 4.86
MM1 pKa = 7.69SKK3 pKa = 9.0RR4 pKa = 11.84TFQPNNRR11 pKa = 11.84RR12 pKa = 11.84RR13 pKa = 11.84AKK15 pKa = 10.04KK16 pKa = 8.86HH17 pKa = 4.36GFRR20 pKa = 11.84ARR22 pKa = 11.84MRR24 pKa = 11.84TRR26 pKa = 11.84AGRR29 pKa = 11.84SILAARR35 pKa = 11.84RR36 pKa = 11.84SKK38 pKa = 10.97GRR40 pKa = 11.84TEE42 pKa = 4.38LSAA45 pKa = 4.86
Molecular weight: 5.27 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
868408 |
37 |
2114 |
319.2 |
34.17 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
13.912 ± 0.062 | 0.484 ± 0.01 |
6.588 ± 0.039 | 5.743 ± 0.039 |
3.089 ± 0.025 | 8.871 ± 0.039 |
1.972 ± 0.023 | 4.507 ± 0.029 |
1.73 ± 0.031 | 9.992 ± 0.048 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.742 ± 0.019 | 1.758 ± 0.021 |
5.521 ± 0.039 | 2.584 ± 0.026 |
7.825 ± 0.055 | 5.253 ± 0.028 |
5.941 ± 0.035 | 9.078 ± 0.04 |
1.506 ± 0.021 | 1.903 ± 0.019 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
