
Polymorphobacter arshaanensis
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Alphaproteobacteria; Sphingomonadales; Sphingosinicellaceae; Polymorphobacter
Average proteome isoelectric point is 6.59
Get precalculated fractions of proteins
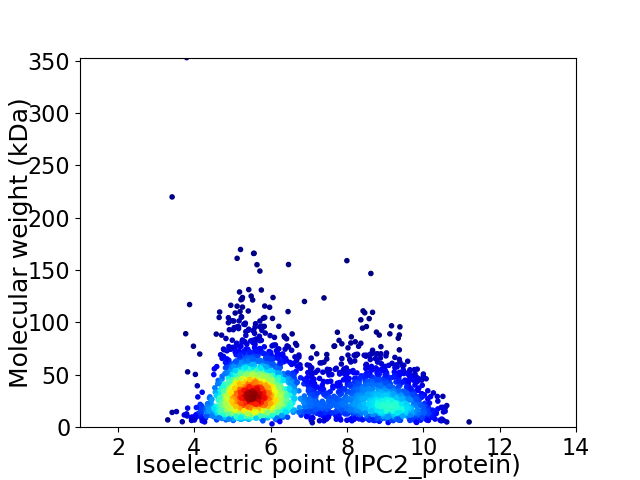
Virtual 2D-PAGE plot for 3035 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A4Y9EQG0|A0A4Y9EQG0_9SPHN Rhodanese-like domain-containing protein OS=Polymorphobacter arshaanensis OX=2511025 GN=EUV02_00200 PE=4 SV=1
MM1 pKa = 6.46TTQTIQVSTAAGLMSALASATGGEE25 pKa = 4.66TIQLAAGNYY34 pKa = 8.67GGVYY38 pKa = 10.05LYY40 pKa = 10.52QYY42 pKa = 11.54NFAQKK47 pKa = 10.14VNIVGGTFSSIQVVSSNGLNFEE69 pKa = 4.1GTTVNFVPDD78 pKa = 3.54ATSTSNSQGVRR89 pKa = 11.84VYY91 pKa = 10.86LSQNINFNNSTVVGGLSVNGVDD113 pKa = 4.36PSAASGDD120 pKa = 3.46ATGNVLGYY128 pKa = 9.61PVGKK132 pKa = 10.05GVNFEE137 pKa = 4.21NSSFCSITNSDD148 pKa = 3.15VSVFAKK154 pKa = 10.66GITITGGHH162 pKa = 7.47DD163 pKa = 3.28YY164 pKa = 11.21LIDD167 pKa = 3.72NNTIHH172 pKa = 7.83DD173 pKa = 4.46LRR175 pKa = 11.84TTPISGSVTSNLTISNNHH193 pKa = 5.54TWDD196 pKa = 3.2SHH198 pKa = 4.52PWQFGTQDD206 pKa = 3.01HH207 pKa = 7.21GDD209 pKa = 4.2RR210 pKa = 11.84IHH212 pKa = 6.25VWTNGTPISGLVIANNYY229 pKa = 9.09LDD231 pKa = 4.17QGAGDD236 pKa = 4.17PMLGIYY242 pKa = 10.41LDD244 pKa = 4.48DD245 pKa = 4.52NGAGMGFTNAVISGNTLIDD264 pKa = 3.44GTGQGVLLEE273 pKa = 4.45NVTGTVSNNTLIWSGTGTQANNSPRR298 pKa = 11.84FDD300 pKa = 3.37INAGSDD306 pKa = 3.62HH307 pKa = 7.5INFTDD312 pKa = 3.21NTGNVSIQTGSTFLNFVRR330 pKa = 11.84QTGMIAEE337 pKa = 4.73DD338 pKa = 3.55ATMSSADD345 pKa = 4.3RR346 pKa = 11.84DD347 pKa = 4.45TITIDD352 pKa = 3.26YY353 pKa = 9.3MVITAHH359 pKa = 6.92NSYY362 pKa = 10.85ALNATTSDD370 pKa = 4.23LYY372 pKa = 10.88FQGGIGDD379 pKa = 4.07FVGTGNTLANHH390 pKa = 6.4IVGSTGNDD398 pKa = 3.04TLTGNGGADD407 pKa = 3.51YY408 pKa = 11.49LEE410 pKa = 4.78GKK412 pKa = 7.95TGNDD416 pKa = 3.16TYY418 pKa = 11.75VVDD421 pKa = 4.27NLSQVIYY428 pKa = 10.59DD429 pKa = 3.52SGGTEE434 pKa = 4.02LVKK437 pKa = 11.04SKK439 pKa = 10.68ISWTLQTGLEE449 pKa = 4.06NLTFIGTGSATLAGNAAPNIIIGGGGADD477 pKa = 3.26RR478 pKa = 11.84LIGNGGADD486 pKa = 3.63TLQGGLGNDD495 pKa = 3.57TYY497 pKa = 11.63VIDD500 pKa = 4.74NLGQKK505 pKa = 8.24ITDD508 pKa = 3.44TGGTDD513 pKa = 3.47TIEE516 pKa = 5.17ASLNWTLQSGIEE528 pKa = 3.97NLTLTGAAVFATGNTSANTLRR549 pKa = 11.84GNGLNNTLDD558 pKa = 3.83GKK560 pKa = 9.2TGADD564 pKa = 3.22SMYY567 pKa = 10.73GGAGDD572 pKa = 5.69DD573 pKa = 4.26IYY575 pKa = 10.98IVDD578 pKa = 3.71NAGDD582 pKa = 3.54KK583 pKa = 10.7VFEE586 pKa = 4.32VVAGIDD592 pKa = 3.11SGGFDD597 pKa = 3.57TVKK600 pKa = 9.88TALASWTMAAGVEE613 pKa = 4.27VLTYY617 pKa = 9.46TGSAAFTGTGNTLNNTISGGAAIDD641 pKa = 3.89TLNGLAGDD649 pKa = 4.76DD650 pKa = 4.1KK651 pKa = 11.67LIGNAGDD658 pKa = 4.06DD659 pKa = 4.56KK660 pKa = 11.46INGGAGSDD668 pKa = 3.65TLTGGTGNDD677 pKa = 2.87TFIFYY682 pKa = 10.53RR683 pKa = 11.84GEE685 pKa = 4.18AYY687 pKa = 10.45SDD689 pKa = 3.98SITDD693 pKa = 3.51FNGRR697 pKa = 11.84ISTVGDD703 pKa = 3.8SIEE706 pKa = 3.82LRR708 pKa = 11.84GWGTGSTFTNIAGTNFWVIHH728 pKa = 7.01DD729 pKa = 4.28GLDD732 pKa = 3.47GYY734 pKa = 10.45EE735 pKa = 4.27EE736 pKa = 4.7YY737 pKa = 10.53ISVTGGVDD745 pKa = 3.2PSDD748 pKa = 3.07WSFTQSVTSVVAPAAAVMPMSVLALDD774 pKa = 3.9TQGVDD779 pKa = 3.64LTSVAATEE787 pKa = 4.18TDD789 pKa = 3.69LVLATSDD796 pKa = 3.81TPDD799 pKa = 3.5LSSVLDD805 pKa = 4.15DD806 pKa = 5.72SVDD809 pKa = 3.63QTDD812 pKa = 3.9MFPDD816 pKa = 4.58DD817 pKa = 3.93SAQAAQQVTEE827 pKa = 4.43DD828 pKa = 4.21AGTFDD833 pKa = 3.94PAVPEE838 pKa = 3.75AAAAQGHH845 pKa = 6.6DD846 pKa = 3.6GGRR849 pKa = 11.84DD850 pKa = 3.98GVWLPFANADD860 pKa = 3.25HH861 pKa = 6.53HH862 pKa = 6.95VPYY865 pKa = 10.52HH866 pKa = 5.77HH867 pKa = 7.39AA868 pKa = 4.16
MM1 pKa = 6.46TTQTIQVSTAAGLMSALASATGGEE25 pKa = 4.66TIQLAAGNYY34 pKa = 8.67GGVYY38 pKa = 10.05LYY40 pKa = 10.52QYY42 pKa = 11.54NFAQKK47 pKa = 10.14VNIVGGTFSSIQVVSSNGLNFEE69 pKa = 4.1GTTVNFVPDD78 pKa = 3.54ATSTSNSQGVRR89 pKa = 11.84VYY91 pKa = 10.86LSQNINFNNSTVVGGLSVNGVDD113 pKa = 4.36PSAASGDD120 pKa = 3.46ATGNVLGYY128 pKa = 9.61PVGKK132 pKa = 10.05GVNFEE137 pKa = 4.21NSSFCSITNSDD148 pKa = 3.15VSVFAKK154 pKa = 10.66GITITGGHH162 pKa = 7.47DD163 pKa = 3.28YY164 pKa = 11.21LIDD167 pKa = 3.72NNTIHH172 pKa = 7.83DD173 pKa = 4.46LRR175 pKa = 11.84TTPISGSVTSNLTISNNHH193 pKa = 5.54TWDD196 pKa = 3.2SHH198 pKa = 4.52PWQFGTQDD206 pKa = 3.01HH207 pKa = 7.21GDD209 pKa = 4.2RR210 pKa = 11.84IHH212 pKa = 6.25VWTNGTPISGLVIANNYY229 pKa = 9.09LDD231 pKa = 4.17QGAGDD236 pKa = 4.17PMLGIYY242 pKa = 10.41LDD244 pKa = 4.48DD245 pKa = 4.52NGAGMGFTNAVISGNTLIDD264 pKa = 3.44GTGQGVLLEE273 pKa = 4.45NVTGTVSNNTLIWSGTGTQANNSPRR298 pKa = 11.84FDD300 pKa = 3.37INAGSDD306 pKa = 3.62HH307 pKa = 7.5INFTDD312 pKa = 3.21NTGNVSIQTGSTFLNFVRR330 pKa = 11.84QTGMIAEE337 pKa = 4.73DD338 pKa = 3.55ATMSSADD345 pKa = 4.3RR346 pKa = 11.84DD347 pKa = 4.45TITIDD352 pKa = 3.26YY353 pKa = 9.3MVITAHH359 pKa = 6.92NSYY362 pKa = 10.85ALNATTSDD370 pKa = 4.23LYY372 pKa = 10.88FQGGIGDD379 pKa = 4.07FVGTGNTLANHH390 pKa = 6.4IVGSTGNDD398 pKa = 3.04TLTGNGGADD407 pKa = 3.51YY408 pKa = 11.49LEE410 pKa = 4.78GKK412 pKa = 7.95TGNDD416 pKa = 3.16TYY418 pKa = 11.75VVDD421 pKa = 4.27NLSQVIYY428 pKa = 10.59DD429 pKa = 3.52SGGTEE434 pKa = 4.02LVKK437 pKa = 11.04SKK439 pKa = 10.68ISWTLQTGLEE449 pKa = 4.06NLTFIGTGSATLAGNAAPNIIIGGGGADD477 pKa = 3.26RR478 pKa = 11.84LIGNGGADD486 pKa = 3.63TLQGGLGNDD495 pKa = 3.57TYY497 pKa = 11.63VIDD500 pKa = 4.74NLGQKK505 pKa = 8.24ITDD508 pKa = 3.44TGGTDD513 pKa = 3.47TIEE516 pKa = 5.17ASLNWTLQSGIEE528 pKa = 3.97NLTLTGAAVFATGNTSANTLRR549 pKa = 11.84GNGLNNTLDD558 pKa = 3.83GKK560 pKa = 9.2TGADD564 pKa = 3.22SMYY567 pKa = 10.73GGAGDD572 pKa = 5.69DD573 pKa = 4.26IYY575 pKa = 10.98IVDD578 pKa = 3.71NAGDD582 pKa = 3.54KK583 pKa = 10.7VFEE586 pKa = 4.32VVAGIDD592 pKa = 3.11SGGFDD597 pKa = 3.57TVKK600 pKa = 9.88TALASWTMAAGVEE613 pKa = 4.27VLTYY617 pKa = 9.46TGSAAFTGTGNTLNNTISGGAAIDD641 pKa = 3.89TLNGLAGDD649 pKa = 4.76DD650 pKa = 4.1KK651 pKa = 11.67LIGNAGDD658 pKa = 4.06DD659 pKa = 4.56KK660 pKa = 11.46INGGAGSDD668 pKa = 3.65TLTGGTGNDD677 pKa = 2.87TFIFYY682 pKa = 10.53RR683 pKa = 11.84GEE685 pKa = 4.18AYY687 pKa = 10.45SDD689 pKa = 3.98SITDD693 pKa = 3.51FNGRR697 pKa = 11.84ISTVGDD703 pKa = 3.8SIEE706 pKa = 3.82LRR708 pKa = 11.84GWGTGSTFTNIAGTNFWVIHH728 pKa = 7.01DD729 pKa = 4.28GLDD732 pKa = 3.47GYY734 pKa = 10.45EE735 pKa = 4.27EE736 pKa = 4.7YY737 pKa = 10.53ISVTGGVDD745 pKa = 3.2PSDD748 pKa = 3.07WSFTQSVTSVVAPAAAVMPMSVLALDD774 pKa = 3.9TQGVDD779 pKa = 3.64LTSVAATEE787 pKa = 4.18TDD789 pKa = 3.69LVLATSDD796 pKa = 3.81TPDD799 pKa = 3.5LSSVLDD805 pKa = 4.15DD806 pKa = 5.72SVDD809 pKa = 3.63QTDD812 pKa = 3.9MFPDD816 pKa = 4.58DD817 pKa = 3.93SAQAAQQVTEE827 pKa = 4.43DD828 pKa = 4.21AGTFDD833 pKa = 3.94PAVPEE838 pKa = 3.75AAAAQGHH845 pKa = 6.6DD846 pKa = 3.6GGRR849 pKa = 11.84DD850 pKa = 3.98GVWLPFANADD860 pKa = 3.25HH861 pKa = 6.53HH862 pKa = 6.95VPYY865 pKa = 10.52HH866 pKa = 5.77HH867 pKa = 7.39AA868 pKa = 4.16
Molecular weight: 89.26 kDa
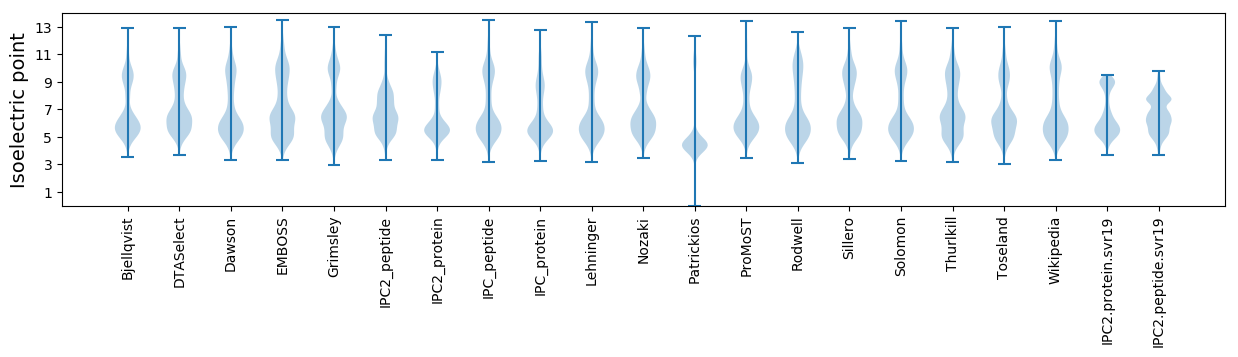
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A4Y9EN67|A0A4Y9EN67_9SPHN Replication-associated recombination protein A OS=Polymorphobacter arshaanensis OX=2511025 GN=EUV02_09930 PE=3 SV=1
MM1 pKa = 7.45KK2 pKa = 9.59RR3 pKa = 11.84TFQPSKK9 pKa = 9.58LVRR12 pKa = 11.84ARR14 pKa = 11.84RR15 pKa = 11.84HH16 pKa = 4.36GFRR19 pKa = 11.84TRR21 pKa = 11.84MATPGGRR28 pKa = 11.84KK29 pKa = 9.01VLANRR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 10.45GRR39 pKa = 11.84AKK41 pKa = 10.69LSAA44 pKa = 3.92
MM1 pKa = 7.45KK2 pKa = 9.59RR3 pKa = 11.84TFQPSKK9 pKa = 9.58LVRR12 pKa = 11.84ARR14 pKa = 11.84RR15 pKa = 11.84HH16 pKa = 4.36GFRR19 pKa = 11.84TRR21 pKa = 11.84MATPGGRR28 pKa = 11.84KK29 pKa = 9.01VLANRR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 10.45GRR39 pKa = 11.84AKK41 pKa = 10.69LSAA44 pKa = 3.92
Molecular weight: 5.02 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
983693 |
28 |
3624 |
324.1 |
34.71 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
14.307 ± 0.065 | 0.763 ± 0.014 |
5.832 ± 0.034 | 4.551 ± 0.042 |
3.605 ± 0.023 | 8.996 ± 0.052 |
2.012 ± 0.022 | 4.946 ± 0.032 |
3.087 ± 0.036 | 9.958 ± 0.05 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.452 ± 0.022 | 2.742 ± 0.035 |
5.372 ± 0.035 | 2.865 ± 0.021 |
6.535 ± 0.057 | 4.958 ± 0.029 |
5.719 ± 0.042 | 7.57 ± 0.035 |
1.468 ± 0.019 | 2.261 ± 0.022 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
