
Escherichia phage Penshu1
Taxonomy: Viruses; Duplodnaviria; Heunggongvirae; Uroviricota; Caudoviricetes; Caudovirales; Autographiviridae; Studiervirinae; Kayfunavirus; unclassified Kayfunavirus
Average proteome isoelectric point is 6.93
Get precalculated fractions of proteins
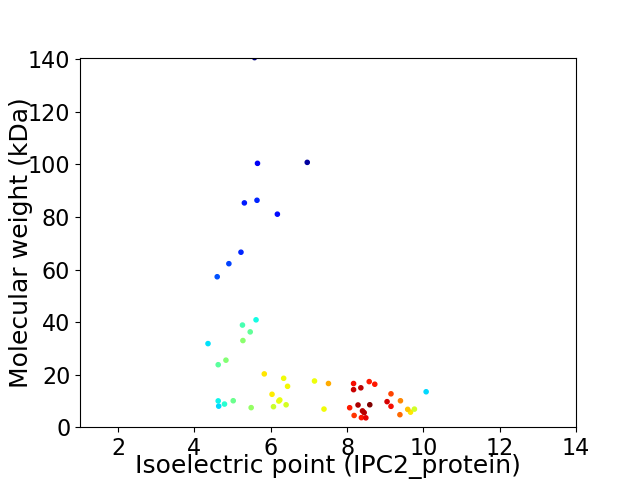
Virtual 2D-PAGE plot for 54 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions

Protein with the lowest isoelectric point:
>tr|A0A5B9NAG1|A0A5B9NAG1_9CAUD Internal virion protein gp15 OS=Escherichia phage Penshu1 OX=2591101 GN=CPT_Penshu1_044 PE=3 SV=1
MM1 pKa = 6.87SQSVYY6 pKa = 11.38AEE8 pKa = 4.16FGVSSNAVTGSVEE21 pKa = 5.06DD22 pKa = 4.16LNEE25 pKa = 4.16HH26 pKa = 5.47QKK28 pKa = 11.61SMLEE32 pKa = 3.55QDD34 pKa = 3.61VAVRR38 pKa = 11.84DD39 pKa = 3.75GDD41 pKa = 3.85DD42 pKa = 4.58AITFKK47 pKa = 10.85QLEE50 pKa = 4.43AEE52 pKa = 4.22SEE54 pKa = 4.16EE55 pKa = 4.02ATEE58 pKa = 3.71EE59 pKa = 4.17DD60 pKa = 4.03EE61 pKa = 5.06NVEE64 pKa = 4.05EE65 pKa = 4.5TEE67 pKa = 4.0GEE69 pKa = 4.0EE70 pKa = 4.22DD71 pKa = 4.13HH72 pKa = 7.29EE73 pKa = 5.84SDD75 pKa = 5.18DD76 pKa = 4.98EE77 pKa = 4.22EE78 pKa = 5.98SEE80 pKa = 4.31TDD82 pKa = 3.5GEE84 pKa = 4.22QPEE87 pKa = 4.71FIEE90 pKa = 5.95LGDD93 pKa = 3.86APKK96 pKa = 10.67EE97 pKa = 3.96LTEE100 pKa = 4.52SVTALDD106 pKa = 3.81EE107 pKa = 4.32NEE109 pKa = 4.15AAFDD113 pKa = 4.13DD114 pKa = 4.55MVSSAVEE121 pKa = 3.96AGKK124 pKa = 8.43VTADD128 pKa = 3.95EE129 pKa = 4.11ITSIKK134 pKa = 10.63AEE136 pKa = 3.97YY137 pKa = 10.49AKK139 pKa = 10.65DD140 pKa = 3.61GKK142 pKa = 11.09LSDD145 pKa = 3.33ASYY148 pKa = 11.61AKK150 pKa = 10.33LQEE153 pKa = 3.93AGYY156 pKa = 7.75TKK158 pKa = 10.52RR159 pKa = 11.84FVDD162 pKa = 3.55SFVRR166 pKa = 11.84GQEE169 pKa = 3.91ALAEE173 pKa = 4.09QYY175 pKa = 10.62AAGVVRR181 pKa = 11.84YY182 pKa = 9.78AGGAEE187 pKa = 3.89QFNRR191 pKa = 11.84ILSHH195 pKa = 6.95LEE197 pKa = 3.78SNDD200 pKa = 2.97PSTRR204 pKa = 11.84EE205 pKa = 3.83ALEE208 pKa = 3.76AAIVRR213 pKa = 11.84KK214 pKa = 10.32DD215 pKa = 3.1IATTKK220 pKa = 10.96ALLNLAGKK228 pKa = 7.93TLGKK232 pKa = 10.41AVGVKK237 pKa = 8.57PQRR240 pKa = 11.84TITAQAKK247 pKa = 9.26PVVAPKK253 pKa = 10.58APQTEE258 pKa = 4.38AFSSKK263 pKa = 10.93ADD265 pKa = 3.78MIKK268 pKa = 10.77AMSDD272 pKa = 2.96PRR274 pKa = 11.84YY275 pKa = 10.34LRR277 pKa = 11.84DD278 pKa = 3.04AKK280 pKa = 10.36YY281 pKa = 9.52TMEE284 pKa = 3.95VRR286 pKa = 11.84AKK288 pKa = 10.22VAASSLL294 pKa = 3.78
MM1 pKa = 6.87SQSVYY6 pKa = 11.38AEE8 pKa = 4.16FGVSSNAVTGSVEE21 pKa = 5.06DD22 pKa = 4.16LNEE25 pKa = 4.16HH26 pKa = 5.47QKK28 pKa = 11.61SMLEE32 pKa = 3.55QDD34 pKa = 3.61VAVRR38 pKa = 11.84DD39 pKa = 3.75GDD41 pKa = 3.85DD42 pKa = 4.58AITFKK47 pKa = 10.85QLEE50 pKa = 4.43AEE52 pKa = 4.22SEE54 pKa = 4.16EE55 pKa = 4.02ATEE58 pKa = 3.71EE59 pKa = 4.17DD60 pKa = 4.03EE61 pKa = 5.06NVEE64 pKa = 4.05EE65 pKa = 4.5TEE67 pKa = 4.0GEE69 pKa = 4.0EE70 pKa = 4.22DD71 pKa = 4.13HH72 pKa = 7.29EE73 pKa = 5.84SDD75 pKa = 5.18DD76 pKa = 4.98EE77 pKa = 4.22EE78 pKa = 5.98SEE80 pKa = 4.31TDD82 pKa = 3.5GEE84 pKa = 4.22QPEE87 pKa = 4.71FIEE90 pKa = 5.95LGDD93 pKa = 3.86APKK96 pKa = 10.67EE97 pKa = 3.96LTEE100 pKa = 4.52SVTALDD106 pKa = 3.81EE107 pKa = 4.32NEE109 pKa = 4.15AAFDD113 pKa = 4.13DD114 pKa = 4.55MVSSAVEE121 pKa = 3.96AGKK124 pKa = 8.43VTADD128 pKa = 3.95EE129 pKa = 4.11ITSIKK134 pKa = 10.63AEE136 pKa = 3.97YY137 pKa = 10.49AKK139 pKa = 10.65DD140 pKa = 3.61GKK142 pKa = 11.09LSDD145 pKa = 3.33ASYY148 pKa = 11.61AKK150 pKa = 10.33LQEE153 pKa = 3.93AGYY156 pKa = 7.75TKK158 pKa = 10.52RR159 pKa = 11.84FVDD162 pKa = 3.55SFVRR166 pKa = 11.84GQEE169 pKa = 3.91ALAEE173 pKa = 4.09QYY175 pKa = 10.62AAGVVRR181 pKa = 11.84YY182 pKa = 9.78AGGAEE187 pKa = 3.89QFNRR191 pKa = 11.84ILSHH195 pKa = 6.95LEE197 pKa = 3.78SNDD200 pKa = 2.97PSTRR204 pKa = 11.84EE205 pKa = 3.83ALEE208 pKa = 3.76AAIVRR213 pKa = 11.84KK214 pKa = 10.32DD215 pKa = 3.1IATTKK220 pKa = 10.96ALLNLAGKK228 pKa = 7.93TLGKK232 pKa = 10.41AVGVKK237 pKa = 8.57PQRR240 pKa = 11.84TITAQAKK247 pKa = 9.26PVVAPKK253 pKa = 10.58APQTEE258 pKa = 4.38AFSSKK263 pKa = 10.93ADD265 pKa = 3.78MIKK268 pKa = 10.77AMSDD272 pKa = 2.96PRR274 pKa = 11.84YY275 pKa = 10.34LRR277 pKa = 11.84DD278 pKa = 3.04AKK280 pKa = 10.36YY281 pKa = 9.52TMEE284 pKa = 3.95VRR286 pKa = 11.84AKK288 pKa = 10.22VAASSLL294 pKa = 3.78
Molecular weight: 31.82 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A5B9N6C4|A0A5B9N6C4_9CAUD SsDNA binding protein OS=Escherichia phage Penshu1 OX=2591101 GN=CPT_Penshu1_016 PE=4 SV=1
MM1 pKa = 7.73VDD3 pKa = 3.64EE4 pKa = 5.21RR5 pKa = 11.84SSTDD9 pKa = 2.49RR10 pKa = 11.84WPQRR14 pKa = 11.84LPLTLIGVNYY24 pKa = 7.29EE25 pKa = 3.8HH26 pKa = 7.63RR27 pKa = 11.84LLGMLISGNLSDD39 pKa = 5.06SLGASGRR46 pKa = 11.84NQARR50 pKa = 11.84VILMSWILPVIFGYY64 pKa = 10.85GLIAYY69 pKa = 9.38VMGRR73 pKa = 11.84DD74 pKa = 3.14INKK77 pKa = 9.24ARR79 pKa = 11.84KK80 pKa = 8.78VYY82 pKa = 9.84KK83 pKa = 10.44LNYY86 pKa = 8.5VRR88 pKa = 11.84LGRR91 pKa = 11.84WTVRR95 pKa = 11.84QPNGRR100 pKa = 11.84FMRR103 pKa = 11.84NLANVWDD110 pKa = 4.02IATLGSKK117 pKa = 10.07LL118 pKa = 3.58
MM1 pKa = 7.73VDD3 pKa = 3.64EE4 pKa = 5.21RR5 pKa = 11.84SSTDD9 pKa = 2.49RR10 pKa = 11.84WPQRR14 pKa = 11.84LPLTLIGVNYY24 pKa = 7.29EE25 pKa = 3.8HH26 pKa = 7.63RR27 pKa = 11.84LLGMLISGNLSDD39 pKa = 5.06SLGASGRR46 pKa = 11.84NQARR50 pKa = 11.84VILMSWILPVIFGYY64 pKa = 10.85GLIAYY69 pKa = 9.38VMGRR73 pKa = 11.84DD74 pKa = 3.14INKK77 pKa = 9.24ARR79 pKa = 11.84KK80 pKa = 8.78VYY82 pKa = 9.84KK83 pKa = 10.44LNYY86 pKa = 8.5VRR88 pKa = 11.84LGRR91 pKa = 11.84WTVRR95 pKa = 11.84QPNGRR100 pKa = 11.84FMRR103 pKa = 11.84NLANVWDD110 pKa = 4.02IATLGSKK117 pKa = 10.07LL118 pKa = 3.58
Molecular weight: 13.49 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
12641 |
30 |
1296 |
234.1 |
26.03 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
8.583 ± 0.437 | 1.068 ± 0.235 |
6.408 ± 0.264 | 6.503 ± 0.34 |
3.52 ± 0.183 | 8.006 ± 0.385 |
1.827 ± 0.198 | 4.778 ± 0.152 |
6.685 ± 0.345 | 8.196 ± 0.282 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.864 ± 0.158 | 4.383 ± 0.224 |
3.789 ± 0.165 | 3.742 ± 0.367 |
5.506 ± 0.163 | 6.329 ± 0.325 |
5.807 ± 0.244 | 7.017 ± 0.261 |
1.456 ± 0.171 | 3.536 ± 0.153 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
