
Pacific flying fox faeces associated gemycircularvirus-2
Taxonomy: Viruses; Monodnaviria; Shotokuvirae; Cressdnaviricota; Repensiviricetes; Geplafuvirales; Genomoviridae; Gemycircularvirus; Gemycircularvirus ptero2; Pteropus associated gemycircularvirus 2
Average proteome isoelectric point is 7.0
Get precalculated fractions of proteins
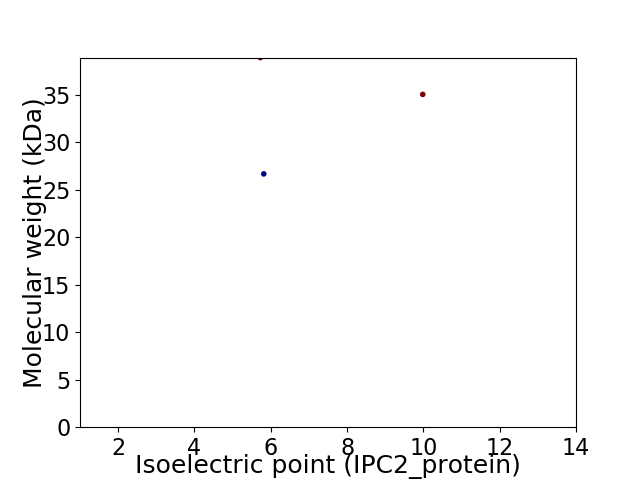
Virtual 2D-PAGE plot for 3 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
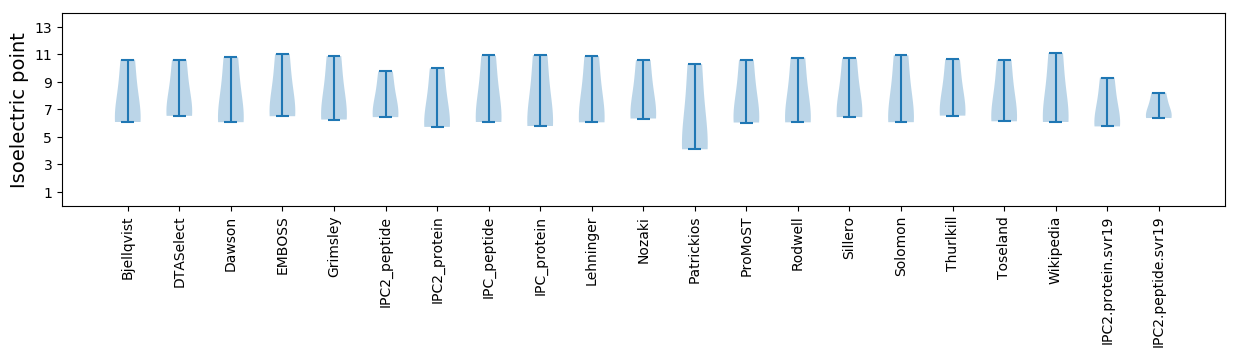
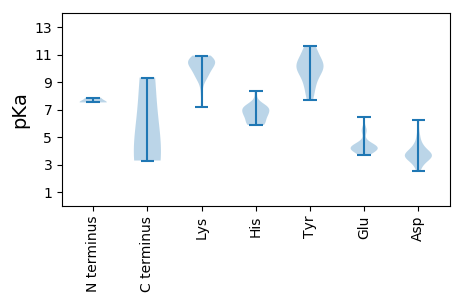
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A140CTL3|A0A140CTL3_9VIRU RepA OS=Pacific flying fox faeces associated gemycircularvirus-2 OX=1795994 PE=3 SV=1
MM1 pKa = 7.52SFYY4 pKa = 10.7FCARR8 pKa = 11.84YY9 pKa = 9.94GLFTYY14 pKa = 7.73SQCADD19 pKa = 3.59LNHH22 pKa = 6.75WSVLDD27 pKa = 3.89LFSGLGAEE35 pKa = 4.62CIIGRR40 pKa = 11.84EE41 pKa = 4.06LHH43 pKa = 6.69EE44 pKa = 5.62DD45 pKa = 3.63GGTHH49 pKa = 6.0LHH51 pKa = 5.94VFADD55 pKa = 4.51FGRR58 pKa = 11.84RR59 pKa = 11.84FRR61 pKa = 11.84SRR63 pKa = 11.84SSKK66 pKa = 10.5IFDD69 pKa = 3.52CEE71 pKa = 3.87GRR73 pKa = 11.84HH74 pKa = 5.88PNVSASRR81 pKa = 11.84GKK83 pKa = 9.64PDD85 pKa = 3.84EE86 pKa = 4.41GWDD89 pKa = 3.65YY90 pKa = 11.6AVKK93 pKa = 10.85DD94 pKa = 3.75GDD96 pKa = 4.04VVAGGLEE103 pKa = 4.06RR104 pKa = 11.84PDD106 pKa = 3.18GVTRR110 pKa = 11.84RR111 pKa = 11.84SGDD114 pKa = 3.41STSDD118 pKa = 3.5QKK120 pKa = 10.77WGEE123 pKa = 3.71IAGAEE128 pKa = 4.23DD129 pKa = 4.33RR130 pKa = 11.84DD131 pKa = 4.16EE132 pKa = 4.4FWRR135 pKa = 11.84LVHH138 pKa = 6.6EE139 pKa = 5.37LDD141 pKa = 3.76PKK143 pKa = 10.92SLVIHH148 pKa = 6.44FPAISKK154 pKa = 9.21YY155 pKa = 10.09CDD157 pKa = 2.66WKK159 pKa = 10.33FSPRR163 pKa = 11.84QVEE166 pKa = 4.27YY167 pKa = 10.58VHH169 pKa = 7.13PINIDD174 pKa = 3.59FVGGEE179 pKa = 4.07VDD181 pKa = 6.26GRR183 pKa = 11.84DD184 pKa = 3.02HH185 pKa = 7.12WLAQAGIGLGGTGKK199 pKa = 10.08SPQMNTWGPRR209 pKa = 11.84FHH211 pKa = 7.18PVGLRR216 pKa = 11.84GMKK219 pKa = 9.75SLCVYY224 pKa = 10.15GKK226 pKa = 10.4SRR228 pKa = 11.84TGKK231 pKa = 7.21TLWARR236 pKa = 11.84SLGRR240 pKa = 11.84HH241 pKa = 6.26IYY243 pKa = 10.61CIGLVSGAEE252 pKa = 4.07CARR255 pKa = 11.84AEE257 pKa = 4.24EE258 pKa = 3.7VDD260 pKa = 3.82YY261 pKa = 11.52AVFDD265 pKa = 5.17DD266 pKa = 3.64IRR268 pKa = 11.84GGIKK272 pKa = 10.04FFHH275 pKa = 7.18AYY277 pKa = 8.84KK278 pKa = 10.42EE279 pKa = 4.32WMGAQAMVSVKK290 pKa = 10.42LLYY293 pKa = 10.06RR294 pKa = 11.84DD295 pKa = 3.67PKK297 pKa = 9.71LVRR300 pKa = 11.84WGKK303 pKa = 9.42PSIWLSNKK311 pKa = 9.94DD312 pKa = 3.65PRR314 pKa = 11.84CDD316 pKa = 3.25MSQEE320 pKa = 3.98DD321 pKa = 4.7VEE323 pKa = 4.31WLEE326 pKa = 4.29DD327 pKa = 3.16NCIFVEE333 pKa = 4.32CNEE336 pKa = 4.45TIFRR340 pKa = 11.84ANTEE344 pKa = 3.92
MM1 pKa = 7.52SFYY4 pKa = 10.7FCARR8 pKa = 11.84YY9 pKa = 9.94GLFTYY14 pKa = 7.73SQCADD19 pKa = 3.59LNHH22 pKa = 6.75WSVLDD27 pKa = 3.89LFSGLGAEE35 pKa = 4.62CIIGRR40 pKa = 11.84EE41 pKa = 4.06LHH43 pKa = 6.69EE44 pKa = 5.62DD45 pKa = 3.63GGTHH49 pKa = 6.0LHH51 pKa = 5.94VFADD55 pKa = 4.51FGRR58 pKa = 11.84RR59 pKa = 11.84FRR61 pKa = 11.84SRR63 pKa = 11.84SSKK66 pKa = 10.5IFDD69 pKa = 3.52CEE71 pKa = 3.87GRR73 pKa = 11.84HH74 pKa = 5.88PNVSASRR81 pKa = 11.84GKK83 pKa = 9.64PDD85 pKa = 3.84EE86 pKa = 4.41GWDD89 pKa = 3.65YY90 pKa = 11.6AVKK93 pKa = 10.85DD94 pKa = 3.75GDD96 pKa = 4.04VVAGGLEE103 pKa = 4.06RR104 pKa = 11.84PDD106 pKa = 3.18GVTRR110 pKa = 11.84RR111 pKa = 11.84SGDD114 pKa = 3.41STSDD118 pKa = 3.5QKK120 pKa = 10.77WGEE123 pKa = 3.71IAGAEE128 pKa = 4.23DD129 pKa = 4.33RR130 pKa = 11.84DD131 pKa = 4.16EE132 pKa = 4.4FWRR135 pKa = 11.84LVHH138 pKa = 6.6EE139 pKa = 5.37LDD141 pKa = 3.76PKK143 pKa = 10.92SLVIHH148 pKa = 6.44FPAISKK154 pKa = 9.21YY155 pKa = 10.09CDD157 pKa = 2.66WKK159 pKa = 10.33FSPRR163 pKa = 11.84QVEE166 pKa = 4.27YY167 pKa = 10.58VHH169 pKa = 7.13PINIDD174 pKa = 3.59FVGGEE179 pKa = 4.07VDD181 pKa = 6.26GRR183 pKa = 11.84DD184 pKa = 3.02HH185 pKa = 7.12WLAQAGIGLGGTGKK199 pKa = 10.08SPQMNTWGPRR209 pKa = 11.84FHH211 pKa = 7.18PVGLRR216 pKa = 11.84GMKK219 pKa = 9.75SLCVYY224 pKa = 10.15GKK226 pKa = 10.4SRR228 pKa = 11.84TGKK231 pKa = 7.21TLWARR236 pKa = 11.84SLGRR240 pKa = 11.84HH241 pKa = 6.26IYY243 pKa = 10.61CIGLVSGAEE252 pKa = 4.07CARR255 pKa = 11.84AEE257 pKa = 4.24EE258 pKa = 3.7VDD260 pKa = 3.82YY261 pKa = 11.52AVFDD265 pKa = 5.17DD266 pKa = 3.64IRR268 pKa = 11.84GGIKK272 pKa = 10.04FFHH275 pKa = 7.18AYY277 pKa = 8.84KK278 pKa = 10.42EE279 pKa = 4.32WMGAQAMVSVKK290 pKa = 10.42LLYY293 pKa = 10.06RR294 pKa = 11.84DD295 pKa = 3.67PKK297 pKa = 9.71LVRR300 pKa = 11.84WGKK303 pKa = 9.42PSIWLSNKK311 pKa = 9.94DD312 pKa = 3.65PRR314 pKa = 11.84CDD316 pKa = 3.25MSQEE320 pKa = 3.98DD321 pKa = 4.7VEE323 pKa = 4.31WLEE326 pKa = 4.29DD327 pKa = 3.16NCIFVEE333 pKa = 4.32CNEE336 pKa = 4.45TIFRR340 pKa = 11.84ANTEE344 pKa = 3.92
Molecular weight: 38.92 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A140CTL2|A0A140CTL2_9VIRU Replication-associated protein OS=Pacific flying fox faeces associated gemycircularvirus-2 OX=1795994 PE=3 SV=1
MM1 pKa = 7.86AYY3 pKa = 9.52PRR5 pKa = 11.84RR6 pKa = 11.84SYY8 pKa = 10.78RR9 pKa = 11.84RR10 pKa = 11.84ASGRR14 pKa = 11.84RR15 pKa = 11.84RR16 pKa = 11.84VSRR19 pKa = 11.84RR20 pKa = 11.84APRR23 pKa = 11.84RR24 pKa = 11.84TKK26 pKa = 8.32RR27 pKa = 11.84TYY29 pKa = 9.12RR30 pKa = 11.84RR31 pKa = 11.84TYY33 pKa = 8.42RR34 pKa = 11.84RR35 pKa = 11.84RR36 pKa = 11.84RR37 pKa = 11.84VGGRR41 pKa = 11.84GGRR44 pKa = 11.84RR45 pKa = 11.84SLLNATSRR53 pKa = 11.84KK54 pKa = 9.16KK55 pKa = 10.63RR56 pKa = 11.84NTMLQFVNTNSTSGLPVTPGPGPYY80 pKa = 9.61VVAGNQGSAFSVFTATAMDD99 pKa = 5.05LYY101 pKa = 11.49ANGNANTIANQAQRR115 pKa = 11.84TSTTCYY121 pKa = 9.27VLGYY125 pKa = 10.01AEE127 pKa = 5.45KK128 pKa = 10.47IRR130 pKa = 11.84IQTSSGLPWFWRR142 pKa = 11.84RR143 pKa = 11.84IVVRR147 pKa = 11.84VRR149 pKa = 11.84DD150 pKa = 4.03PLFVTYY156 pKa = 8.58ATNDD160 pKa = 3.38TPTYY164 pKa = 8.14SANMSYY170 pKa = 11.39VDD172 pKa = 3.42TTTNGMQRR180 pKa = 11.84AYY182 pKa = 9.98INQTINAANNTITGVQGVLFRR203 pKa = 11.84GTVGRR208 pKa = 11.84DD209 pKa = 2.66WTDD212 pKa = 2.53IQTAIVDD219 pKa = 3.85TTRR222 pKa = 11.84VDD224 pKa = 3.72LVSDD228 pKa = 3.17TRR230 pKa = 11.84MTIRR234 pKa = 11.84SGNANGTVKK243 pKa = 10.38DD244 pKa = 3.68VNMWHH249 pKa = 7.53PYY251 pKa = 9.03RR252 pKa = 11.84RR253 pKa = 11.84NLVYY257 pKa = 10.77DD258 pKa = 4.09DD259 pKa = 5.63DD260 pKa = 4.35EE261 pKa = 6.47LGEE264 pKa = 4.25QEE266 pKa = 4.07TANYY270 pKa = 10.15VSTEE274 pKa = 4.03SKK276 pKa = 10.5QGGGDD281 pKa = 3.18MHH283 pKa = 8.32VIDD286 pKa = 5.07IFTPGTGAGASDD298 pKa = 4.5LLQLTSTSTLYY309 pKa = 9.69WHH311 pKa = 7.1EE312 pKa = 4.2KK313 pKa = 9.32
MM1 pKa = 7.86AYY3 pKa = 9.52PRR5 pKa = 11.84RR6 pKa = 11.84SYY8 pKa = 10.78RR9 pKa = 11.84RR10 pKa = 11.84ASGRR14 pKa = 11.84RR15 pKa = 11.84RR16 pKa = 11.84VSRR19 pKa = 11.84RR20 pKa = 11.84APRR23 pKa = 11.84RR24 pKa = 11.84TKK26 pKa = 8.32RR27 pKa = 11.84TYY29 pKa = 9.12RR30 pKa = 11.84RR31 pKa = 11.84TYY33 pKa = 8.42RR34 pKa = 11.84RR35 pKa = 11.84RR36 pKa = 11.84RR37 pKa = 11.84VGGRR41 pKa = 11.84GGRR44 pKa = 11.84RR45 pKa = 11.84SLLNATSRR53 pKa = 11.84KK54 pKa = 9.16KK55 pKa = 10.63RR56 pKa = 11.84NTMLQFVNTNSTSGLPVTPGPGPYY80 pKa = 9.61VVAGNQGSAFSVFTATAMDD99 pKa = 5.05LYY101 pKa = 11.49ANGNANTIANQAQRR115 pKa = 11.84TSTTCYY121 pKa = 9.27VLGYY125 pKa = 10.01AEE127 pKa = 5.45KK128 pKa = 10.47IRR130 pKa = 11.84IQTSSGLPWFWRR142 pKa = 11.84RR143 pKa = 11.84IVVRR147 pKa = 11.84VRR149 pKa = 11.84DD150 pKa = 4.03PLFVTYY156 pKa = 8.58ATNDD160 pKa = 3.38TPTYY164 pKa = 8.14SANMSYY170 pKa = 11.39VDD172 pKa = 3.42TTTNGMQRR180 pKa = 11.84AYY182 pKa = 9.98INQTINAANNTITGVQGVLFRR203 pKa = 11.84GTVGRR208 pKa = 11.84DD209 pKa = 2.66WTDD212 pKa = 2.53IQTAIVDD219 pKa = 3.85TTRR222 pKa = 11.84VDD224 pKa = 3.72LVSDD228 pKa = 3.17TRR230 pKa = 11.84MTIRR234 pKa = 11.84SGNANGTVKK243 pKa = 10.38DD244 pKa = 3.68VNMWHH249 pKa = 7.53PYY251 pKa = 9.03RR252 pKa = 11.84RR253 pKa = 11.84NLVYY257 pKa = 10.77DD258 pKa = 4.09DD259 pKa = 5.63DD260 pKa = 4.35EE261 pKa = 6.47LGEE264 pKa = 4.25QEE266 pKa = 4.07TANYY270 pKa = 10.15VSTEE274 pKa = 4.03SKK276 pKa = 10.5QGGGDD281 pKa = 3.18MHH283 pKa = 8.32VIDD286 pKa = 5.07IFTPGTGAGASDD298 pKa = 4.5LLQLTSTSTLYY309 pKa = 9.69WHH311 pKa = 7.1EE312 pKa = 4.2KK313 pKa = 9.32
Molecular weight: 35.04 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
896 |
239 |
344 |
298.7 |
33.55 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.25 ± 0.559 | 2.009 ± 0.79 |
6.92 ± 0.829 | 4.464 ± 1.208 |
4.353 ± 0.98 | 10.491 ± 0.946 |
2.79 ± 0.854 | 4.353 ± 0.238 |
3.571 ± 0.71 | 6.027 ± 0.43 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.786 ± 0.408 | 3.683 ± 1.394 |
3.906 ± 0.258 | 2.679 ± 0.53 |
9.152 ± 1.227 | 6.92 ± 0.101 |
6.696 ± 2.803 | 7.478 ± 0.39 |
2.79 ± 0.548 | 3.683 ± 0.673 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
