
Ensete ventricosum (Abyssinian banana) (Musa ensete)
Taxonomy: cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliopsida; Mesangiospermae; Liliopsida; Petrosaviidae; commelinids; Zingiberales; Musaceae; Ensete
Average proteome isoelectric point is 7.4
Get precalculated fractions of proteins
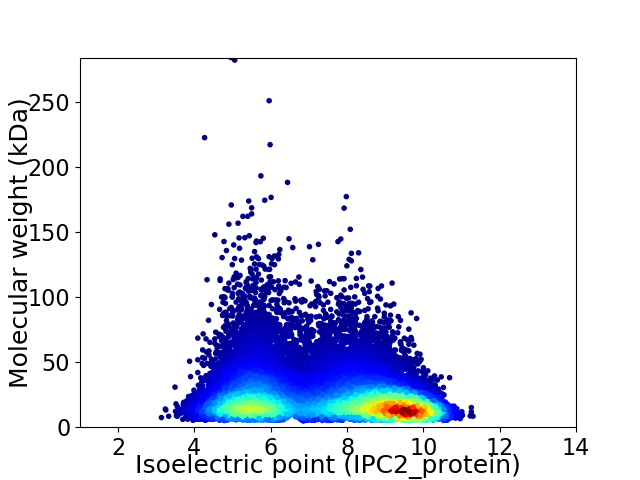
Virtual 2D-PAGE plot for 58382 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
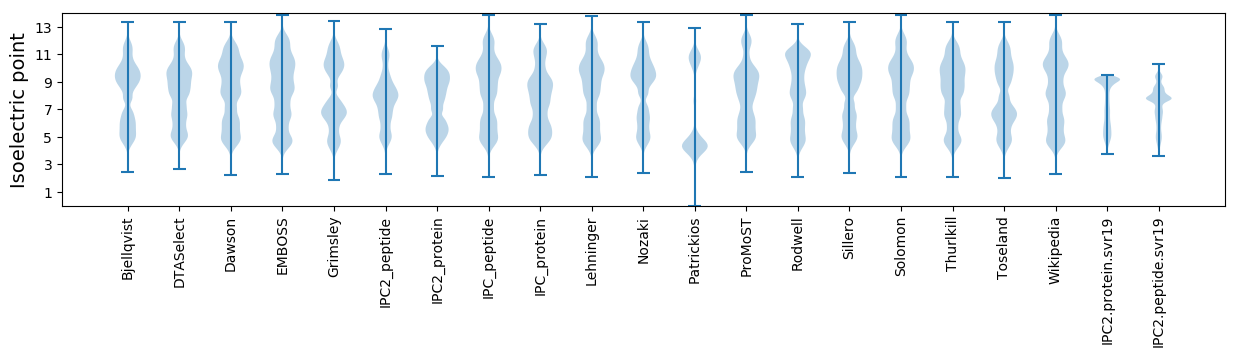
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A444F8W1|A0A444F8W1_ENSVE Uncharacterized protein OS=Ensete ventricosum OX=4639 GN=GW17_00016912 PE=4 SV=1
MM1 pKa = 7.77ASLLLSPPSLADD13 pKa = 3.73PNATTMPTPSGAIQRR28 pKa = 11.84EE29 pKa = 3.85MDD31 pKa = 3.5QQILATVLPDD41 pKa = 3.53SSSNTQPFLTSPTGKK56 pKa = 8.5YY57 pKa = 8.13TGYY60 pKa = 11.03LLRR63 pKa = 11.84HH64 pKa = 6.4KK65 pKa = 8.72STPDD69 pKa = 2.9SGGYY73 pKa = 10.54GSDD76 pKa = 3.02FCYY79 pKa = 10.39IQIQDD84 pKa = 3.96TVSGDD89 pKa = 4.05SVWEE93 pKa = 4.32SEE95 pKa = 4.39CEE97 pKa = 4.06PVSSANACTLVFSDD111 pKa = 4.88AGLAVLDD118 pKa = 4.92GSQSVWDD125 pKa = 4.0TGASNGNNFPATLEE139 pKa = 4.03LADD142 pKa = 4.86LGDD145 pKa = 3.57MTVADD150 pKa = 4.49KK151 pKa = 11.25DD152 pKa = 4.08GEE154 pKa = 4.63LVWKK158 pKa = 10.38ASNDD162 pKa = 3.04PRR164 pKa = 11.84VNQGCVSPDD173 pKa = 3.45LGSGSPTFVGGDD185 pKa = 3.58DD186 pKa = 4.14NSPLGQQLPPPPPYY200 pKa = 10.23LSEE203 pKa = 4.26GALPFAPASPPVVGDD218 pKa = 3.94DD219 pKa = 5.71DD220 pKa = 4.01ITSGQAPLLAPATAPFAAPTSGDD243 pKa = 3.17EE244 pKa = 4.25NIPYY248 pKa = 9.12NQQPTSYY255 pKa = 9.45PSNGAFGQQQEE266 pKa = 4.35QQQSAFHH273 pKa = 6.45GLHH276 pKa = 6.19GVNQPLLDD284 pKa = 3.87NNAYY288 pKa = 10.48DD289 pKa = 3.73SGCSRR294 pKa = 11.84KK295 pKa = 9.7EE296 pKa = 3.7GLVGILLAVVGHH308 pKa = 6.3LVLRR312 pKa = 11.84GLL314 pKa = 4.08
MM1 pKa = 7.77ASLLLSPPSLADD13 pKa = 3.73PNATTMPTPSGAIQRR28 pKa = 11.84EE29 pKa = 3.85MDD31 pKa = 3.5QQILATVLPDD41 pKa = 3.53SSSNTQPFLTSPTGKK56 pKa = 8.5YY57 pKa = 8.13TGYY60 pKa = 11.03LLRR63 pKa = 11.84HH64 pKa = 6.4KK65 pKa = 8.72STPDD69 pKa = 2.9SGGYY73 pKa = 10.54GSDD76 pKa = 3.02FCYY79 pKa = 10.39IQIQDD84 pKa = 3.96TVSGDD89 pKa = 4.05SVWEE93 pKa = 4.32SEE95 pKa = 4.39CEE97 pKa = 4.06PVSSANACTLVFSDD111 pKa = 4.88AGLAVLDD118 pKa = 4.92GSQSVWDD125 pKa = 4.0TGASNGNNFPATLEE139 pKa = 4.03LADD142 pKa = 4.86LGDD145 pKa = 3.57MTVADD150 pKa = 4.49KK151 pKa = 11.25DD152 pKa = 4.08GEE154 pKa = 4.63LVWKK158 pKa = 10.38ASNDD162 pKa = 3.04PRR164 pKa = 11.84VNQGCVSPDD173 pKa = 3.45LGSGSPTFVGGDD185 pKa = 3.58DD186 pKa = 4.14NSPLGQQLPPPPPYY200 pKa = 10.23LSEE203 pKa = 4.26GALPFAPASPPVVGDD218 pKa = 3.94DD219 pKa = 5.71DD220 pKa = 4.01ITSGQAPLLAPATAPFAAPTSGDD243 pKa = 3.17EE244 pKa = 4.25NIPYY248 pKa = 9.12NQQPTSYY255 pKa = 9.45PSNGAFGQQQEE266 pKa = 4.35QQQSAFHH273 pKa = 6.45GLHH276 pKa = 6.19GVNQPLLDD284 pKa = 3.87NNAYY288 pKa = 10.48DD289 pKa = 3.73SGCSRR294 pKa = 11.84KK295 pKa = 9.7EE296 pKa = 3.7GLVGILLAVVGHH308 pKa = 6.3LVLRR312 pKa = 11.84GLL314 pKa = 4.08
Molecular weight: 32.58 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A444DM74|A0A444DM74_ENSVE Uncharacterized protein (Fragment) OS=Ensete ventricosum OX=4639 GN=GW17_00037851 PE=4 SV=1
MM1 pKa = 7.59GISFRR6 pKa = 11.84VQRR9 pKa = 11.84VPISNAALPPPFSLLGSSRR28 pKa = 11.84GQRR31 pKa = 11.84RR32 pKa = 11.84FKK34 pKa = 10.85HH35 pKa = 5.26FVPSIVLSSLSPLLRR50 pKa = 11.84LSLARR55 pKa = 11.84SHH57 pKa = 6.18IRR59 pKa = 11.84PRR61 pKa = 11.84HH62 pKa = 5.05RR63 pKa = 11.84RR64 pKa = 11.84RR65 pKa = 11.84LWMASRR71 pKa = 11.84TGII74 pKa = 4.04
MM1 pKa = 7.59GISFRR6 pKa = 11.84VQRR9 pKa = 11.84VPISNAALPPPFSLLGSSRR28 pKa = 11.84GQRR31 pKa = 11.84RR32 pKa = 11.84FKK34 pKa = 10.85HH35 pKa = 5.26FVPSIVLSSLSPLLRR50 pKa = 11.84LSLARR55 pKa = 11.84SHH57 pKa = 6.18IRR59 pKa = 11.84PRR61 pKa = 11.84HH62 pKa = 5.05RR63 pKa = 11.84RR64 pKa = 11.84RR65 pKa = 11.84LWMASRR71 pKa = 11.84TGII74 pKa = 4.04
Molecular weight: 8.43 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
13105813 |
49 |
3236 |
224.5 |
24.84 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
8.07 ± 0.012 | 2.112 ± 0.006 |
4.941 ± 0.008 | 6.016 ± 0.013 |
3.733 ± 0.009 | 7.319 ± 0.012 |
2.563 ± 0.006 | 4.283 ± 0.009 |
4.885 ± 0.011 | 9.576 ± 0.015 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.356 ± 0.006 | 3.211 ± 0.008 |
5.899 ± 0.014 | 3.306 ± 0.009 |
7.386 ± 0.012 | 9.161 ± 0.014 |
4.817 ± 0.007 | 6.633 ± 0.01 |
1.354 ± 0.004 | 2.376 ± 0.006 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
