
Stipagrostis associaed virus
Taxonomy: Viruses; unclassified viruses; unclassified DNA viruses
Average proteome isoelectric point is 7.71
Get precalculated fractions of proteins
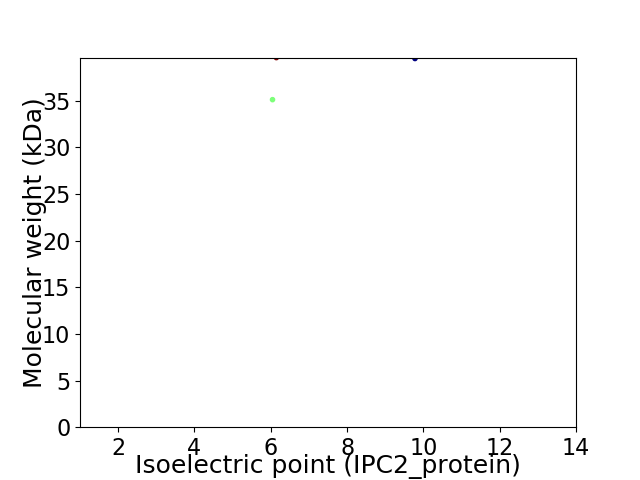
Virtual 2D-PAGE plot for 2 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A345BKA4|A0A345BKA4_9VIRU Putative capsid protein OS=Stipagrostis associaed virus OX=2282646 PE=4 SV=1
MM1 pKa = 7.0YY2 pKa = 9.99RR3 pKa = 11.84IRR5 pKa = 11.84GQALLLTYY13 pKa = 10.42SQTSLDD19 pKa = 4.05QNDD22 pKa = 3.85LAAFHH27 pKa = 7.07HH28 pKa = 6.15EE29 pKa = 4.05QLPISYY35 pKa = 10.05YY36 pKa = 10.51LLANEE41 pKa = 4.18THH43 pKa = 6.25QDD45 pKa = 3.12NGRR48 pKa = 11.84HH49 pKa = 3.76YY50 pKa = 10.28HH51 pKa = 6.39VYY53 pKa = 10.94LEE55 pKa = 4.25TEE57 pKa = 3.83KK58 pKa = 11.0RR59 pKa = 11.84FDD61 pKa = 3.18ITNVRR66 pKa = 11.84RR67 pKa = 11.84FDD69 pKa = 3.95FGGEE73 pKa = 4.1HH74 pKa = 7.56PNTQVPRR81 pKa = 11.84NRR83 pKa = 11.84EE84 pKa = 3.56ATINYY89 pKa = 7.36CKK91 pKa = 10.48KK92 pKa = 10.7EE93 pKa = 3.61GDD95 pKa = 3.48YY96 pKa = 10.98WEE98 pKa = 4.92LGRR101 pKa = 11.84PATNASRR108 pKa = 11.84QQHH111 pKa = 5.6LCEE114 pKa = 4.07EE115 pKa = 4.83TTFSTGDD122 pKa = 3.17CRR124 pKa = 11.84NFEE127 pKa = 5.06KK128 pKa = 10.79EE129 pKa = 3.83IDD131 pKa = 4.04WIDD134 pKa = 3.19HH135 pKa = 6.68CITNKK140 pKa = 9.87IQFGYY145 pKa = 9.59CARR148 pKa = 11.84IWALHH153 pKa = 5.69NIKK156 pKa = 9.64PLKK159 pKa = 10.54YY160 pKa = 8.53ITNDD164 pKa = 3.43SIFNGVVTHH173 pKa = 6.16PTLMLPIQPDD183 pKa = 3.59PAKK186 pKa = 8.83ATVVIGPTGVGKK198 pKa = 7.48TVWAKK203 pKa = 10.55RR204 pKa = 11.84NSIKK208 pKa = 10.25PALLIRR214 pKa = 11.84HH215 pKa = 6.52LEE217 pKa = 4.04DD218 pKa = 4.02LKK220 pKa = 10.59TVDD223 pKa = 2.77WSYY226 pKa = 11.83LKK228 pKa = 10.07TIIFDD233 pKa = 3.93DD234 pKa = 3.46MLFRR238 pKa = 11.84HH239 pKa = 6.38WPLQTQIHH247 pKa = 6.15LLDD250 pKa = 3.62WDD252 pKa = 4.05EE253 pKa = 5.19DD254 pKa = 4.26SSIHH258 pKa = 6.37ARR260 pKa = 11.84YY261 pKa = 10.29GNITIPANTHH271 pKa = 6.93KK272 pKa = 10.57IFTCNEE278 pKa = 3.75DD279 pKa = 3.33PFIDD283 pKa = 3.55HH284 pKa = 6.48EE285 pKa = 4.88AIFRR289 pKa = 11.84RR290 pKa = 11.84IHH292 pKa = 5.84VYY294 pKa = 10.2RR295 pKa = 11.84LSVTDD300 pKa = 3.55
MM1 pKa = 7.0YY2 pKa = 9.99RR3 pKa = 11.84IRR5 pKa = 11.84GQALLLTYY13 pKa = 10.42SQTSLDD19 pKa = 4.05QNDD22 pKa = 3.85LAAFHH27 pKa = 7.07HH28 pKa = 6.15EE29 pKa = 4.05QLPISYY35 pKa = 10.05YY36 pKa = 10.51LLANEE41 pKa = 4.18THH43 pKa = 6.25QDD45 pKa = 3.12NGRR48 pKa = 11.84HH49 pKa = 3.76YY50 pKa = 10.28HH51 pKa = 6.39VYY53 pKa = 10.94LEE55 pKa = 4.25TEE57 pKa = 3.83KK58 pKa = 11.0RR59 pKa = 11.84FDD61 pKa = 3.18ITNVRR66 pKa = 11.84RR67 pKa = 11.84FDD69 pKa = 3.95FGGEE73 pKa = 4.1HH74 pKa = 7.56PNTQVPRR81 pKa = 11.84NRR83 pKa = 11.84EE84 pKa = 3.56ATINYY89 pKa = 7.36CKK91 pKa = 10.48KK92 pKa = 10.7EE93 pKa = 3.61GDD95 pKa = 3.48YY96 pKa = 10.98WEE98 pKa = 4.92LGRR101 pKa = 11.84PATNASRR108 pKa = 11.84QQHH111 pKa = 5.6LCEE114 pKa = 4.07EE115 pKa = 4.83TTFSTGDD122 pKa = 3.17CRR124 pKa = 11.84NFEE127 pKa = 5.06KK128 pKa = 10.79EE129 pKa = 3.83IDD131 pKa = 4.04WIDD134 pKa = 3.19HH135 pKa = 6.68CITNKK140 pKa = 9.87IQFGYY145 pKa = 9.59CARR148 pKa = 11.84IWALHH153 pKa = 5.69NIKK156 pKa = 9.64PLKK159 pKa = 10.54YY160 pKa = 8.53ITNDD164 pKa = 3.43SIFNGVVTHH173 pKa = 6.16PTLMLPIQPDD183 pKa = 3.59PAKK186 pKa = 8.83ATVVIGPTGVGKK198 pKa = 7.48TVWAKK203 pKa = 10.55RR204 pKa = 11.84NSIKK208 pKa = 10.25PALLIRR214 pKa = 11.84HH215 pKa = 6.52LEE217 pKa = 4.04DD218 pKa = 4.02LKK220 pKa = 10.59TVDD223 pKa = 2.77WSYY226 pKa = 11.83LKK228 pKa = 10.07TIIFDD233 pKa = 3.93DD234 pKa = 3.46MLFRR238 pKa = 11.84HH239 pKa = 6.38WPLQTQIHH247 pKa = 6.15LLDD250 pKa = 3.62WDD252 pKa = 4.05EE253 pKa = 5.19DD254 pKa = 4.26SSIHH258 pKa = 6.37ARR260 pKa = 11.84YY261 pKa = 10.29GNITIPANTHH271 pKa = 6.93KK272 pKa = 10.57IFTCNEE278 pKa = 3.75DD279 pKa = 3.33PFIDD283 pKa = 3.55HH284 pKa = 6.48EE285 pKa = 4.88AIFRR289 pKa = 11.84RR290 pKa = 11.84IHH292 pKa = 5.84VYY294 pKa = 10.2RR295 pKa = 11.84LSVTDD300 pKa = 3.55
Molecular weight: 35.11 kDa
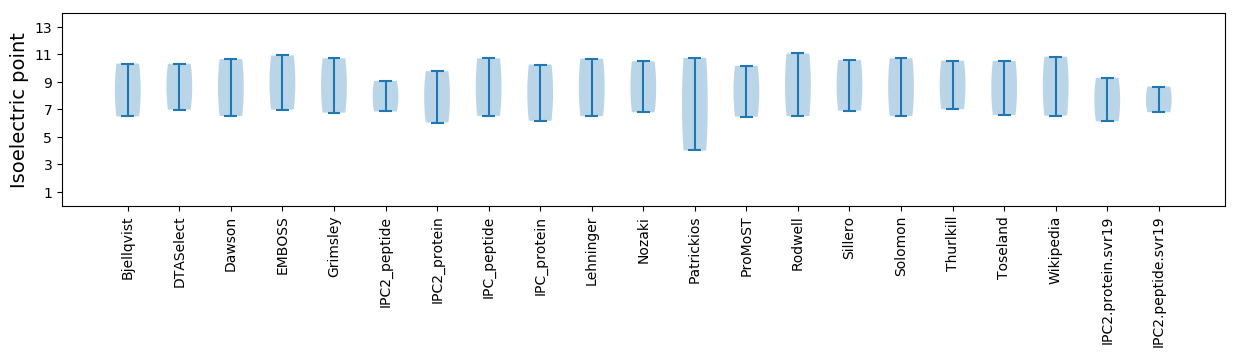
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A345BKA4|A0A345BKA4_9VIRU Putative capsid protein OS=Stipagrostis associaed virus OX=2282646 PE=4 SV=1
MM1 pKa = 7.97PSTITRR7 pKa = 11.84RR8 pKa = 11.84NGVSSYY14 pKa = 10.47RR15 pKa = 11.84PRR17 pKa = 11.84RR18 pKa = 11.84MFGPIGLRR26 pKa = 11.84AMAHH30 pKa = 5.77MGDD33 pKa = 3.72MALKK37 pKa = 9.96RR38 pKa = 11.84ASRR41 pKa = 11.84SYY43 pKa = 11.08KK44 pKa = 9.82SFGRR48 pKa = 11.84RR49 pKa = 11.84GKK51 pKa = 10.07GSSMGKK57 pKa = 9.93SNSKK61 pKa = 9.47MDD63 pKa = 3.63SAPLYY68 pKa = 8.05GTTYY72 pKa = 8.78TSKK75 pKa = 9.84VSYY78 pKa = 10.02RR79 pKa = 11.84KK80 pKa = 9.87RR81 pKa = 11.84GVSRR85 pKa = 11.84SRR87 pKa = 11.84SKK89 pKa = 10.43FGKK92 pKa = 9.82RR93 pKa = 11.84KK94 pKa = 9.21FGKK97 pKa = 9.85FLHH100 pKa = 6.33QSMKK104 pKa = 10.52LQNAQNTLAGNAFGLTTTAGSQAWSSLDD132 pKa = 3.22LCNGVEE138 pKa = 4.7LYY140 pKa = 10.7QILRR144 pKa = 11.84TQLPPSASTATMRR157 pKa = 11.84DD158 pKa = 3.0FTMALKK164 pKa = 10.65NFNLKK169 pKa = 9.65WYY171 pKa = 6.91ITNVGTTNVEE181 pKa = 3.72VEE183 pKa = 4.79IYY185 pKa = 10.23HH186 pKa = 6.06IMPRR190 pKa = 11.84RR191 pKa = 11.84DD192 pKa = 3.43VTCKK196 pKa = 10.06EE197 pKa = 3.92MSIATPSPSNGNIFANYY214 pKa = 9.57VDD216 pKa = 3.71SQYY219 pKa = 11.36PSDD222 pKa = 4.61ALGDD226 pKa = 3.91PDD228 pKa = 4.59GDD230 pKa = 3.65AVPTINSLGWTPFLNRR246 pKa = 11.84NLMRR250 pKa = 11.84QFIVKK255 pKa = 10.16KK256 pKa = 9.9VRR258 pKa = 11.84TVKK261 pKa = 9.56MAPADD266 pKa = 3.58TYY268 pKa = 9.7MEE270 pKa = 4.87RR271 pKa = 11.84DD272 pKa = 3.5SVGQKK277 pKa = 9.01MLNASRR283 pKa = 11.84LGLEE287 pKa = 4.08IFSNGATTSYY297 pKa = 11.48NDD299 pKa = 3.21VQKK302 pKa = 10.48WYY304 pKa = 11.03LKK306 pKa = 10.47GLSQIIFIKK315 pKa = 10.67LRR317 pKa = 11.84GFPNTTSGSPVSSINIAWEE336 pKa = 3.96VTTTSKK342 pKa = 10.91VIQTRR347 pKa = 11.84TGSDD351 pKa = 3.08ALIGNAA357 pKa = 4.43
MM1 pKa = 7.97PSTITRR7 pKa = 11.84RR8 pKa = 11.84NGVSSYY14 pKa = 10.47RR15 pKa = 11.84PRR17 pKa = 11.84RR18 pKa = 11.84MFGPIGLRR26 pKa = 11.84AMAHH30 pKa = 5.77MGDD33 pKa = 3.72MALKK37 pKa = 9.96RR38 pKa = 11.84ASRR41 pKa = 11.84SYY43 pKa = 11.08KK44 pKa = 9.82SFGRR48 pKa = 11.84RR49 pKa = 11.84GKK51 pKa = 10.07GSSMGKK57 pKa = 9.93SNSKK61 pKa = 9.47MDD63 pKa = 3.63SAPLYY68 pKa = 8.05GTTYY72 pKa = 8.78TSKK75 pKa = 9.84VSYY78 pKa = 10.02RR79 pKa = 11.84KK80 pKa = 9.87RR81 pKa = 11.84GVSRR85 pKa = 11.84SRR87 pKa = 11.84SKK89 pKa = 10.43FGKK92 pKa = 9.82RR93 pKa = 11.84KK94 pKa = 9.21FGKK97 pKa = 9.85FLHH100 pKa = 6.33QSMKK104 pKa = 10.52LQNAQNTLAGNAFGLTTTAGSQAWSSLDD132 pKa = 3.22LCNGVEE138 pKa = 4.7LYY140 pKa = 10.7QILRR144 pKa = 11.84TQLPPSASTATMRR157 pKa = 11.84DD158 pKa = 3.0FTMALKK164 pKa = 10.65NFNLKK169 pKa = 9.65WYY171 pKa = 6.91ITNVGTTNVEE181 pKa = 3.72VEE183 pKa = 4.79IYY185 pKa = 10.23HH186 pKa = 6.06IMPRR190 pKa = 11.84RR191 pKa = 11.84DD192 pKa = 3.43VTCKK196 pKa = 10.06EE197 pKa = 3.92MSIATPSPSNGNIFANYY214 pKa = 9.57VDD216 pKa = 3.71SQYY219 pKa = 11.36PSDD222 pKa = 4.61ALGDD226 pKa = 3.91PDD228 pKa = 4.59GDD230 pKa = 3.65AVPTINSLGWTPFLNRR246 pKa = 11.84NLMRR250 pKa = 11.84QFIVKK255 pKa = 10.16KK256 pKa = 9.9VRR258 pKa = 11.84TVKK261 pKa = 9.56MAPADD266 pKa = 3.58TYY268 pKa = 9.7MEE270 pKa = 4.87RR271 pKa = 11.84DD272 pKa = 3.5SVGQKK277 pKa = 9.01MLNASRR283 pKa = 11.84LGLEE287 pKa = 4.08IFSNGATTSYY297 pKa = 11.48NDD299 pKa = 3.21VQKK302 pKa = 10.48WYY304 pKa = 11.03LKK306 pKa = 10.47GLSQIIFIKK315 pKa = 10.67LRR317 pKa = 11.84GFPNTTSGSPVSSINIAWEE336 pKa = 3.96VTTTSKK342 pKa = 10.91VIQTRR347 pKa = 11.84TGSDD351 pKa = 3.08ALIGNAA357 pKa = 4.43
Molecular weight: 39.51 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
657 |
300 |
357 |
328.5 |
37.31 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.088 ± 0.454 | 1.218 ± 0.471 |
5.327 ± 1.007 | 3.501 ± 1.103 |
4.11 ± 0.135 | 6.393 ± 1.239 |
3.044 ± 1.578 | 6.697 ± 1.185 |
5.632 ± 0.581 | 7.61 ± 0.435 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.892 ± 1.138 | 5.936 ± 0.162 |
4.566 ± 0.06 | 3.653 ± 0.209 |
6.849 ± 0.31 | 7.61 ± 2.373 |
8.524 ± 0.114 | 4.566 ± 0.341 |
1.826 ± 0.305 | 3.957 ± 0.226 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
