
Methanoplanus limicola DSM 2279
Taxonomy: cellular organisms; Archaea; Euryarchaeota; Stenosarchaea group; Methanomicrobia; Methanomicrobiales; Methanomicrobiaceae; Methanoplanus; Methanoplanus limicola
Average proteome isoelectric point is 5.74
Get precalculated fractions of proteins
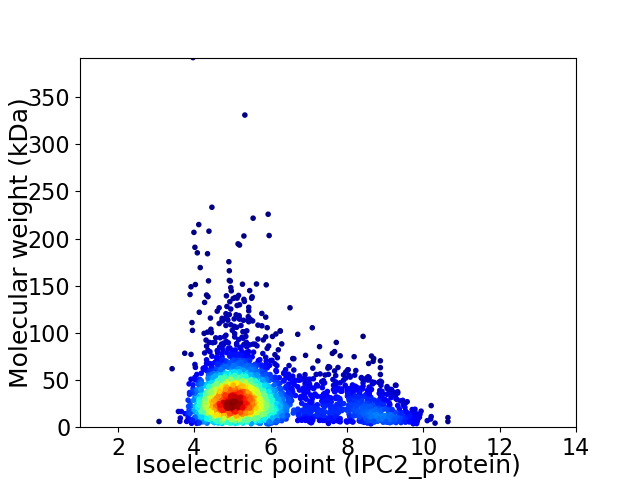
Virtual 2D-PAGE plot for 2928 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|H1Z156|H1Z156_9EURY Putative nickel-responsive regulator OS=Methanoplanus limicola DSM 2279 OX=937775 GN=Metlim_1213 PE=3 SV=1
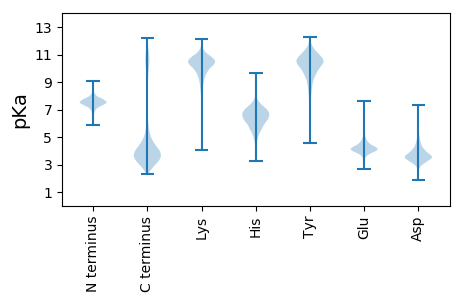
MM1 pKa = 7.76LFFTPVTASPSSGDD15 pKa = 3.58LTDD18 pKa = 3.74AGYY21 pKa = 10.85VALEE25 pKa = 4.33SGDD28 pKa = 3.34NRR30 pKa = 11.84MAKK33 pKa = 9.02TLFTRR38 pKa = 11.84ALDD41 pKa = 3.59TGGQNSPLAWAGLGKK56 pKa = 10.38ALWNDD61 pKa = 3.11SSSLNEE67 pKa = 3.85ASYY70 pKa = 9.08TAFNRR75 pKa = 11.84SLEE78 pKa = 4.25CVNASSEE85 pKa = 3.92DD86 pKa = 3.44WNAVFMVFFNEE97 pKa = 4.06PVNDD101 pKa = 3.8FEE103 pKa = 4.77LAYY106 pKa = 10.75SAGLKK111 pKa = 10.34ALEE114 pKa = 5.0ADD116 pKa = 4.34PEE118 pKa = 5.07DD119 pKa = 4.1DD120 pKa = 4.24VAWNYY125 pKa = 8.63FGCVIDD131 pKa = 5.71GLAHH135 pKa = 6.18NNSDD139 pKa = 3.81ADD141 pKa = 3.97LNEE144 pKa = 4.37PFDD147 pKa = 4.11AFKK150 pKa = 11.04KK151 pKa = 10.68AMSLNPTALYY161 pKa = 9.9SGNVAYY167 pKa = 10.38YY168 pKa = 10.69AIVTGNYY175 pKa = 9.43SYY177 pKa = 11.36AVEE180 pKa = 4.18ITDD183 pKa = 3.79QAVEE187 pKa = 4.34YY188 pKa = 9.69YY189 pKa = 10.86SPDD192 pKa = 3.12EE193 pKa = 4.1WSKK196 pKa = 10.28GTLFFLKK203 pKa = 10.46ALALAEE209 pKa = 4.31TGSSEE214 pKa = 3.95EE215 pKa = 3.72ALEE218 pKa = 4.37NIEE221 pKa = 5.0LSRR224 pKa = 11.84EE225 pKa = 4.13AYY227 pKa = 10.51NSDD230 pKa = 4.27EE231 pKa = 4.18SFEE234 pKa = 4.07WDD236 pKa = 3.42EE237 pKa = 4.0YY238 pKa = 11.28FYY240 pKa = 10.95ATDD243 pKa = 3.6GMVSAIALNDD253 pKa = 3.66LGRR256 pKa = 11.84SDD258 pKa = 4.03EE259 pKa = 4.25SAEE262 pKa = 3.51ILARR266 pKa = 11.84IDD268 pKa = 3.29TDD270 pKa = 3.84KK271 pKa = 11.44LDD273 pKa = 5.78DD274 pKa = 3.97YY275 pKa = 11.08MQGLLFTPEE284 pKa = 4.52RR285 pKa = 11.84YY286 pKa = 8.79MDD288 pKa = 4.42LYY290 pKa = 11.32GVALSGLGRR299 pKa = 11.84TDD301 pKa = 3.69DD302 pKa = 3.63AATAFEE308 pKa = 5.41KK309 pKa = 10.85SLEE312 pKa = 3.96YY313 pKa = 10.98NIDD316 pKa = 3.72YY317 pKa = 10.73IPAKK321 pKa = 10.48NHH323 pKa = 6.82LDD325 pKa = 3.49NLNQKK330 pKa = 9.5ISQQ333 pKa = 3.47
MM1 pKa = 7.76LFFTPVTASPSSGDD15 pKa = 3.58LTDD18 pKa = 3.74AGYY21 pKa = 10.85VALEE25 pKa = 4.33SGDD28 pKa = 3.34NRR30 pKa = 11.84MAKK33 pKa = 9.02TLFTRR38 pKa = 11.84ALDD41 pKa = 3.59TGGQNSPLAWAGLGKK56 pKa = 10.38ALWNDD61 pKa = 3.11SSSLNEE67 pKa = 3.85ASYY70 pKa = 9.08TAFNRR75 pKa = 11.84SLEE78 pKa = 4.25CVNASSEE85 pKa = 3.92DD86 pKa = 3.44WNAVFMVFFNEE97 pKa = 4.06PVNDD101 pKa = 3.8FEE103 pKa = 4.77LAYY106 pKa = 10.75SAGLKK111 pKa = 10.34ALEE114 pKa = 5.0ADD116 pKa = 4.34PEE118 pKa = 5.07DD119 pKa = 4.1DD120 pKa = 4.24VAWNYY125 pKa = 8.63FGCVIDD131 pKa = 5.71GLAHH135 pKa = 6.18NNSDD139 pKa = 3.81ADD141 pKa = 3.97LNEE144 pKa = 4.37PFDD147 pKa = 4.11AFKK150 pKa = 11.04KK151 pKa = 10.68AMSLNPTALYY161 pKa = 9.9SGNVAYY167 pKa = 10.38YY168 pKa = 10.69AIVTGNYY175 pKa = 9.43SYY177 pKa = 11.36AVEE180 pKa = 4.18ITDD183 pKa = 3.79QAVEE187 pKa = 4.34YY188 pKa = 9.69YY189 pKa = 10.86SPDD192 pKa = 3.12EE193 pKa = 4.1WSKK196 pKa = 10.28GTLFFLKK203 pKa = 10.46ALALAEE209 pKa = 4.31TGSSEE214 pKa = 3.95EE215 pKa = 3.72ALEE218 pKa = 4.37NIEE221 pKa = 5.0LSRR224 pKa = 11.84EE225 pKa = 4.13AYY227 pKa = 10.51NSDD230 pKa = 4.27EE231 pKa = 4.18SFEE234 pKa = 4.07WDD236 pKa = 3.42EE237 pKa = 4.0YY238 pKa = 11.28FYY240 pKa = 10.95ATDD243 pKa = 3.6GMVSAIALNDD253 pKa = 3.66LGRR256 pKa = 11.84SDD258 pKa = 4.03EE259 pKa = 4.25SAEE262 pKa = 3.51ILARR266 pKa = 11.84IDD268 pKa = 3.29TDD270 pKa = 3.84KK271 pKa = 11.44LDD273 pKa = 5.78DD274 pKa = 3.97YY275 pKa = 11.08MQGLLFTPEE284 pKa = 4.52RR285 pKa = 11.84YY286 pKa = 8.79MDD288 pKa = 4.42LYY290 pKa = 11.32GVALSGLGRR299 pKa = 11.84TDD301 pKa = 3.69DD302 pKa = 3.63AATAFEE308 pKa = 5.41KK309 pKa = 10.85SLEE312 pKa = 3.96YY313 pKa = 10.98NIDD316 pKa = 3.72YY317 pKa = 10.73IPAKK321 pKa = 10.48NHH323 pKa = 6.82LDD325 pKa = 3.49NLNQKK330 pKa = 9.5ISQQ333 pKa = 3.47
Molecular weight: 36.71 kDa
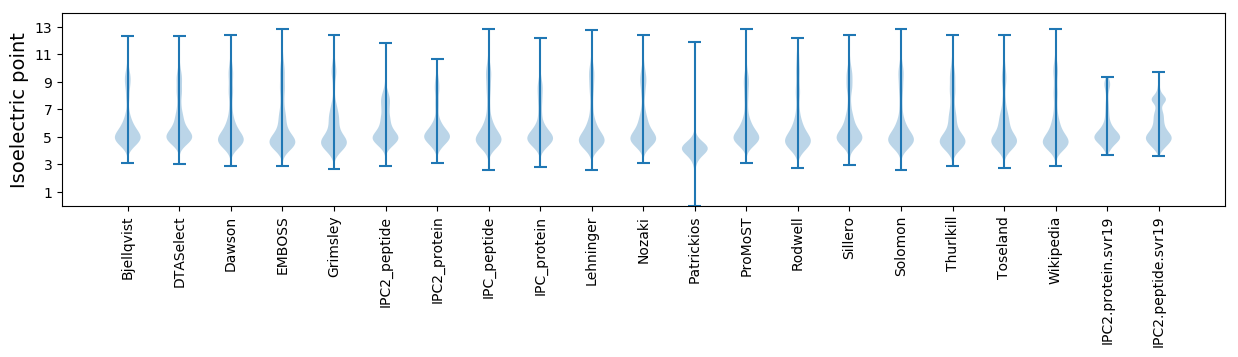
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|H1YWU4|H1YWU4_9EURY 1-(5-phosphoribosyl)-5-amino-4-imidazole-carboxylate (AIR) carboxylase OS=Methanoplanus limicola DSM 2279 OX=937775 GN=Metlim_2800 PE=4 SV=1
MM1 pKa = 7.47AAKK4 pKa = 9.99KK5 pKa = 10.2SSKK8 pKa = 9.27TAFKK12 pKa = 9.93KK13 pKa = 10.23IAMEE17 pKa = 5.05RR18 pKa = 11.84IDD20 pKa = 4.96ILFKK24 pKa = 10.08NAGRR28 pKa = 11.84EE29 pKa = 3.96FSNNPEE35 pKa = 3.76LSNRR39 pKa = 11.84YY40 pKa = 8.72VDD42 pKa = 3.23IARR45 pKa = 11.84RR46 pKa = 11.84IAMRR50 pKa = 11.84QRR52 pKa = 11.84VRR54 pKa = 11.84IPTEE58 pKa = 3.84YY59 pKa = 10.17KK60 pKa = 10.19RR61 pKa = 11.84KK62 pKa = 9.34FCRR65 pKa = 11.84NCHH68 pKa = 5.71SYY70 pKa = 10.87AVPGEE75 pKa = 4.05NMSVRR80 pKa = 11.84IHH82 pKa = 6.53RR83 pKa = 11.84GKK85 pKa = 10.71VIYY88 pKa = 10.15RR89 pKa = 11.84CSQCGYY95 pKa = 9.86IRR97 pKa = 11.84RR98 pKa = 11.84IPINNGDD105 pKa = 3.52RR106 pKa = 11.84KK107 pKa = 10.63
MM1 pKa = 7.47AAKK4 pKa = 9.99KK5 pKa = 10.2SSKK8 pKa = 9.27TAFKK12 pKa = 9.93KK13 pKa = 10.23IAMEE17 pKa = 5.05RR18 pKa = 11.84IDD20 pKa = 4.96ILFKK24 pKa = 10.08NAGRR28 pKa = 11.84EE29 pKa = 3.96FSNNPEE35 pKa = 3.76LSNRR39 pKa = 11.84YY40 pKa = 8.72VDD42 pKa = 3.23IARR45 pKa = 11.84RR46 pKa = 11.84IAMRR50 pKa = 11.84QRR52 pKa = 11.84VRR54 pKa = 11.84IPTEE58 pKa = 3.84YY59 pKa = 10.17KK60 pKa = 10.19RR61 pKa = 11.84KK62 pKa = 9.34FCRR65 pKa = 11.84NCHH68 pKa = 5.71SYY70 pKa = 10.87AVPGEE75 pKa = 4.05NMSVRR80 pKa = 11.84IHH82 pKa = 6.53RR83 pKa = 11.84GKK85 pKa = 10.71VIYY88 pKa = 10.15RR89 pKa = 11.84CSQCGYY95 pKa = 9.86IRR97 pKa = 11.84RR98 pKa = 11.84IPINNGDD105 pKa = 3.52RR106 pKa = 11.84KK107 pKa = 10.63
Molecular weight: 12.6 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
906721 |
32 |
3686 |
309.7 |
34.51 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.868 ± 0.048 | 1.32 ± 0.022 |
6.119 ± 0.038 | 7.74 ± 0.056 |
4.17 ± 0.036 | 7.49 ± 0.044 |
1.609 ± 0.018 | 8.532 ± 0.054 |
6.311 ± 0.053 | 8.562 ± 0.049 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.525 ± 0.026 | 4.562 ± 0.051 |
3.948 ± 0.03 | 2.08 ± 0.022 |
4.711 ± 0.045 | 7.164 ± 0.05 |
5.106 ± 0.055 | 6.508 ± 0.042 |
0.906 ± 0.018 | 3.767 ± 0.034 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
