
Nakamurella panacisegetis
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Actinobacteria; Actinomycetia; Nakamurellales; Nakamurellaceae; Nakamurella
Average proteome isoelectric point is 6.43
Get precalculated fractions of proteins
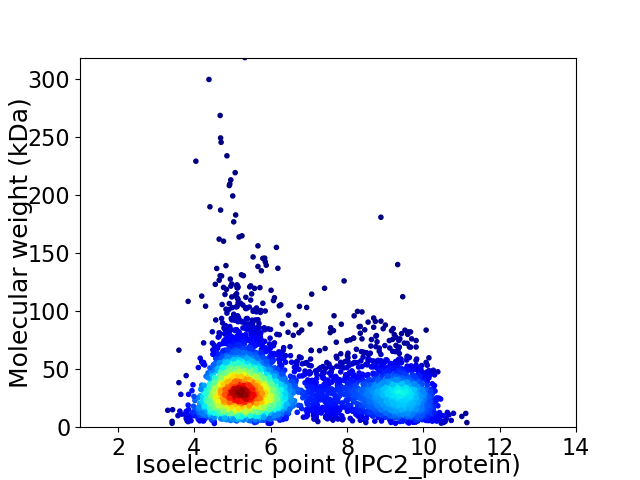
Virtual 2D-PAGE plot for 4387 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
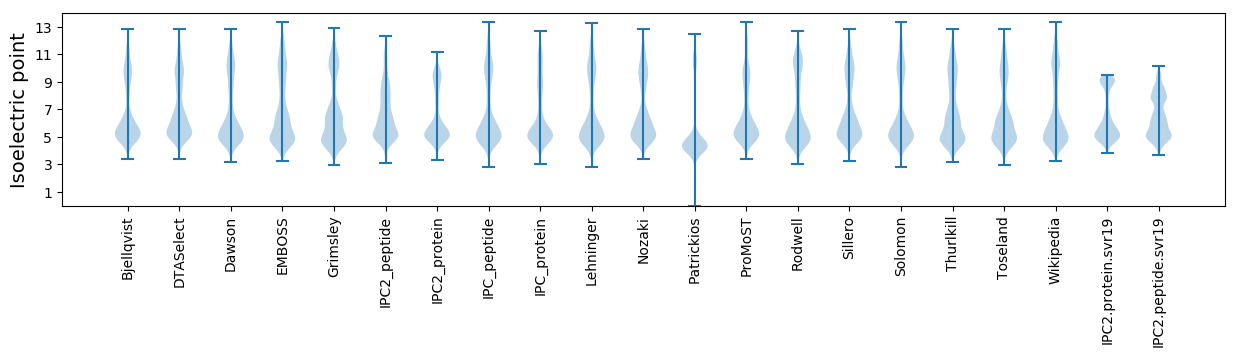
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1H0MGR0|A0A1H0MGR0_9ACTN Uncharacterized protein OS=Nakamurella panacisegetis OX=1090615 GN=SAMN04515671_2009 PE=3 SV=1
MM1 pKa = 7.53EE2 pKa = 6.26FDD4 pKa = 3.99TGPDD8 pKa = 3.44VQPGHH13 pKa = 7.51DD14 pKa = 3.4ISISGSSGSTDD25 pKa = 3.35LGAATYY31 pKa = 10.75DD32 pKa = 3.57SDD34 pKa = 4.27HH35 pKa = 7.38DD36 pKa = 4.19GVADD40 pKa = 3.73SVIVIEE46 pKa = 5.26DD47 pKa = 3.2DD48 pKa = 3.35HH49 pKa = 8.37EE50 pKa = 4.36YY51 pKa = 11.12VITDD55 pKa = 3.56SDD57 pKa = 4.11HH58 pKa = 7.49DD59 pKa = 3.72GHH61 pKa = 7.38ADD63 pKa = 3.54SIHH66 pKa = 7.07AYY68 pKa = 10.17DD69 pKa = 3.84EE70 pKa = 4.6NGNQVDD76 pKa = 4.62PKK78 pKa = 9.96TGAPIDD84 pKa = 3.92GSGDD88 pKa = 3.27GTTGGSSDD96 pKa = 3.52GTTGTDD102 pKa = 3.06TGASTTDD109 pKa = 3.14HH110 pKa = 6.68TGSSTTGASTDD121 pKa = 3.46GTQTGSSSDD130 pKa = 3.59AGSDD134 pKa = 3.2ITVTGSDD141 pKa = 3.72GSAHH145 pKa = 6.9SIGTPTVDD153 pKa = 4.44LDD155 pKa = 4.03HH156 pKa = 7.59DD157 pKa = 4.92GSPDD161 pKa = 3.23TAVVHH166 pKa = 6.33NSDD169 pKa = 3.87GSVTGYY175 pKa = 8.75TDD177 pKa = 3.78RR178 pKa = 11.84DD179 pKa = 3.65GDD181 pKa = 3.81GHH183 pKa = 7.4ADD185 pKa = 3.55QITQIGADD193 pKa = 3.84GKK195 pKa = 10.59VVIAVADD202 pKa = 3.91NNGGWTITTTGHH214 pKa = 6.96LDD216 pKa = 3.05ASGNLVPDD224 pKa = 4.12GTPNPDD230 pKa = 3.4ASLDD234 pKa = 3.76PADD237 pKa = 3.95TAGVTSGHH245 pKa = 5.31TTTSGTATGASDD257 pKa = 3.59HH258 pKa = 5.79TTSGQTTSEE267 pKa = 3.97PTGDD271 pKa = 3.3GRR273 pKa = 11.84TVAEE277 pKa = 4.58PVPNSAPISGNEE289 pKa = 3.74PDD291 pKa = 3.84GDD293 pKa = 3.7ITFSEE298 pKa = 4.5DD299 pKa = 3.42GKK301 pKa = 10.72DD302 pKa = 3.44YY303 pKa = 11.43DD304 pKa = 4.79LGHH307 pKa = 7.23PTTDD311 pKa = 3.64LDD313 pKa = 4.86GDD315 pKa = 4.1GTPDD319 pKa = 3.19TVVTHH324 pKa = 6.83LADD327 pKa = 3.77GTVVGYY333 pKa = 9.39TDD335 pKa = 3.46TDD337 pKa = 3.71GDD339 pKa = 3.92GTADD343 pKa = 3.9QITQISAGGQVVIGVSDD360 pKa = 3.5GHH362 pKa = 6.68GGWTQAATGHH372 pKa = 6.09MGADD376 pKa = 3.6GKK378 pKa = 10.39FVPDD382 pKa = 3.77SSAAATTGG390 pKa = 3.1
MM1 pKa = 7.53EE2 pKa = 6.26FDD4 pKa = 3.99TGPDD8 pKa = 3.44VQPGHH13 pKa = 7.51DD14 pKa = 3.4ISISGSSGSTDD25 pKa = 3.35LGAATYY31 pKa = 10.75DD32 pKa = 3.57SDD34 pKa = 4.27HH35 pKa = 7.38DD36 pKa = 4.19GVADD40 pKa = 3.73SVIVIEE46 pKa = 5.26DD47 pKa = 3.2DD48 pKa = 3.35HH49 pKa = 8.37EE50 pKa = 4.36YY51 pKa = 11.12VITDD55 pKa = 3.56SDD57 pKa = 4.11HH58 pKa = 7.49DD59 pKa = 3.72GHH61 pKa = 7.38ADD63 pKa = 3.54SIHH66 pKa = 7.07AYY68 pKa = 10.17DD69 pKa = 3.84EE70 pKa = 4.6NGNQVDD76 pKa = 4.62PKK78 pKa = 9.96TGAPIDD84 pKa = 3.92GSGDD88 pKa = 3.27GTTGGSSDD96 pKa = 3.52GTTGTDD102 pKa = 3.06TGASTTDD109 pKa = 3.14HH110 pKa = 6.68TGSSTTGASTDD121 pKa = 3.46GTQTGSSSDD130 pKa = 3.59AGSDD134 pKa = 3.2ITVTGSDD141 pKa = 3.72GSAHH145 pKa = 6.9SIGTPTVDD153 pKa = 4.44LDD155 pKa = 4.03HH156 pKa = 7.59DD157 pKa = 4.92GSPDD161 pKa = 3.23TAVVHH166 pKa = 6.33NSDD169 pKa = 3.87GSVTGYY175 pKa = 8.75TDD177 pKa = 3.78RR178 pKa = 11.84DD179 pKa = 3.65GDD181 pKa = 3.81GHH183 pKa = 7.4ADD185 pKa = 3.55QITQIGADD193 pKa = 3.84GKK195 pKa = 10.59VVIAVADD202 pKa = 3.91NNGGWTITTTGHH214 pKa = 6.96LDD216 pKa = 3.05ASGNLVPDD224 pKa = 4.12GTPNPDD230 pKa = 3.4ASLDD234 pKa = 3.76PADD237 pKa = 3.95TAGVTSGHH245 pKa = 5.31TTTSGTATGASDD257 pKa = 3.59HH258 pKa = 5.79TTSGQTTSEE267 pKa = 3.97PTGDD271 pKa = 3.3GRR273 pKa = 11.84TVAEE277 pKa = 4.58PVPNSAPISGNEE289 pKa = 3.74PDD291 pKa = 3.84GDD293 pKa = 3.7ITFSEE298 pKa = 4.5DD299 pKa = 3.42GKK301 pKa = 10.72DD302 pKa = 3.44YY303 pKa = 11.43DD304 pKa = 4.79LGHH307 pKa = 7.23PTTDD311 pKa = 3.64LDD313 pKa = 4.86GDD315 pKa = 4.1GTPDD319 pKa = 3.19TVVTHH324 pKa = 6.83LADD327 pKa = 3.77GTVVGYY333 pKa = 9.39TDD335 pKa = 3.46TDD337 pKa = 3.71GDD339 pKa = 3.92GTADD343 pKa = 3.9QITQISAGGQVVIGVSDD360 pKa = 3.5GHH362 pKa = 6.68GGWTQAATGHH372 pKa = 6.09MGADD376 pKa = 3.6GKK378 pKa = 10.39FVPDD382 pKa = 3.77SSAAATTGG390 pKa = 3.1
Molecular weight: 38.36 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1H0NTZ7|A0A1H0NTZ7_9ACTN Isochorismate synthase OS=Nakamurella panacisegetis OX=1090615 GN=SAMN04515671_2480 PE=3 SV=1
MM1 pKa = 7.4GSVIKK6 pKa = 10.42KK7 pKa = 8.47RR8 pKa = 11.84RR9 pKa = 11.84KK10 pKa = 9.22RR11 pKa = 11.84MAKK14 pKa = 9.41KK15 pKa = 9.87KK16 pKa = 9.7HH17 pKa = 5.84RR18 pKa = 11.84KK19 pKa = 8.7LLRR22 pKa = 11.84KK23 pKa = 7.81TRR25 pKa = 11.84VARR28 pKa = 11.84RR29 pKa = 11.84KK30 pKa = 9.68AGKK33 pKa = 9.73
MM1 pKa = 7.4GSVIKK6 pKa = 10.42KK7 pKa = 8.47RR8 pKa = 11.84RR9 pKa = 11.84KK10 pKa = 9.22RR11 pKa = 11.84MAKK14 pKa = 9.41KK15 pKa = 9.87KK16 pKa = 9.7HH17 pKa = 5.84RR18 pKa = 11.84KK19 pKa = 8.7LLRR22 pKa = 11.84KK23 pKa = 7.81TRR25 pKa = 11.84VARR28 pKa = 11.84RR29 pKa = 11.84KK30 pKa = 9.68AGKK33 pKa = 9.73
Molecular weight: 4.0 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1470634 |
29 |
3217 |
335.2 |
35.59 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
13.449 ± 0.049 | 0.693 ± 0.01 |
5.99 ± 0.032 | 4.527 ± 0.044 |
2.851 ± 0.024 | 9.317 ± 0.036 |
2.06 ± 0.019 | 4.174 ± 0.022 |
2.02 ± 0.028 | 9.927 ± 0.049 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.827 ± 0.015 | 2.082 ± 0.021 |
5.937 ± 0.033 | 2.909 ± 0.018 |
7.172 ± 0.05 | 6.058 ± 0.035 |
6.654 ± 0.052 | 8.971 ± 0.035 |
1.426 ± 0.017 | 1.956 ± 0.02 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
