
Mobiluncus curtisii (strain ATCC 43063 / DSM 2711 / V125) (Falcivibrio vaginalis)
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Actinobacteria; Actinomycetia; Actinomycetales; Actinomycetaceae; Mobiluncus; Mobiluncus curtisii
Average proteome isoelectric point is 6.28
Get precalculated fractions of proteins
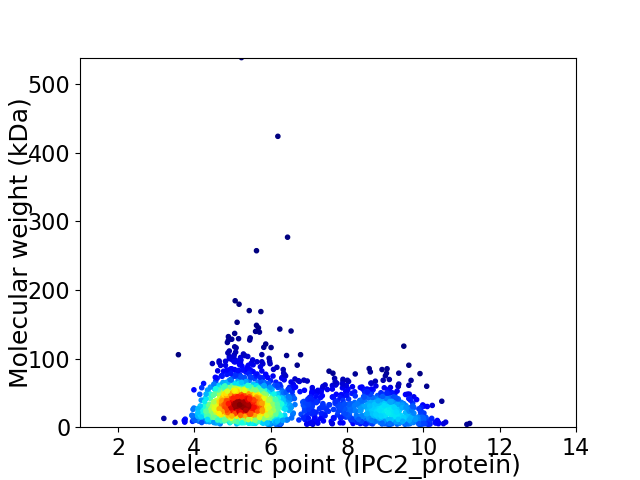
Virtual 2D-PAGE plot for 1904 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|D6ZFU9|D6ZFU9_MOBCV Transcriptional regulator IclR family C-terminal domain protein OS=Mobiluncus curtisii (strain ATCC 43063 / DSM 2711 / V125) OX=548479 GN=HMPREF0573_11188 PE=4 SV=1
MM1 pKa = 7.19YY2 pKa = 9.75AQNPALFGKK11 pKa = 10.16QPATLLTVKK20 pKa = 10.25NGKK23 pKa = 8.79DD24 pKa = 2.84RR25 pKa = 11.84HH26 pKa = 5.31MSEE29 pKa = 5.01AAPQLLTSEE38 pKa = 5.64DD39 pKa = 3.29IFSAQFPATKK49 pKa = 10.13FRR51 pKa = 11.84DD52 pKa = 4.43GYY54 pKa = 10.67DD55 pKa = 3.03QNQIDD60 pKa = 4.67DD61 pKa = 4.11YY62 pKa = 10.39LDD64 pKa = 3.29EE65 pKa = 4.38VVRR68 pKa = 11.84VLSYY72 pKa = 11.35YY73 pKa = 10.02EE74 pKa = 4.37ALNASPEE81 pKa = 4.15AEE83 pKa = 3.87VDD85 pKa = 3.38LAYY88 pKa = 9.33ITVRR92 pKa = 11.84GRR94 pKa = 11.84DD95 pKa = 3.45VRR97 pKa = 11.84EE98 pKa = 3.31VDD100 pKa = 3.83FDD102 pKa = 3.77YY103 pKa = 11.3TRR105 pKa = 11.84MRR107 pKa = 11.84VGYY110 pKa = 9.8DD111 pKa = 2.92QDD113 pKa = 4.8AVDD116 pKa = 5.52DD117 pKa = 4.27YY118 pKa = 11.24LDD120 pKa = 3.46QVAATLEE127 pKa = 4.44AYY129 pKa = 7.77EE130 pKa = 4.45KK131 pKa = 10.77LYY133 pKa = 10.72GIPPSDD139 pKa = 3.23QRR141 pKa = 11.84YY142 pKa = 8.28QVLQVDD148 pKa = 5.16GAALEE153 pKa = 4.51AEE155 pKa = 4.48AAEE158 pKa = 4.15QAAASGEE165 pKa = 4.36LPSSSAPNEE174 pKa = 4.15DD175 pKa = 3.61AAVTPIEE182 pKa = 4.49IAAEE186 pKa = 4.26TPPSQPAEE194 pKa = 3.95QTVASAAPLPVTPAATPSEE213 pKa = 4.14DD214 pKa = 3.65LGVSSSILGTPTGSIPVTQPVPFEE238 pKa = 3.84QAVLTNHH245 pKa = 6.78EE246 pKa = 4.79DD247 pKa = 3.58GSPAGHH253 pKa = 6.48EE254 pKa = 4.22FDD256 pKa = 3.9ATSTASSPLFPPLNAAMSPQSQMPEE281 pKa = 3.8TGTNASPVEE290 pKa = 4.56SIPEE294 pKa = 3.76QNFASAALPEE304 pKa = 4.68TEE306 pKa = 4.36TPIAPSVAASGEE318 pKa = 4.16TAPAVPLPEE327 pKa = 5.67PDD329 pKa = 4.6DD330 pKa = 3.62EE331 pKa = 4.5WGEE334 pKa = 4.29VPQYY338 pKa = 10.56PVDD341 pKa = 3.74APEE344 pKa = 4.43PQTPDD349 pKa = 2.48ATAAQTYY356 pKa = 9.51SEE358 pKa = 4.81PGYY361 pKa = 10.82DD362 pKa = 3.2PYY364 pKa = 11.36QPYY367 pKa = 10.39EE368 pKa = 4.03YY369 pKa = 10.27DD370 pKa = 4.8AYY372 pKa = 10.1DD373 pKa = 3.59QPMSTAPAEE382 pKa = 4.29YY383 pKa = 9.62QGSSEE388 pKa = 4.92PYY390 pKa = 9.31PEE392 pKa = 4.96APTEE396 pKa = 3.95AGYY399 pKa = 9.57PSPYY403 pKa = 10.26GAATPEE409 pKa = 4.82GYY411 pKa = 10.09DD412 pKa = 3.55YY413 pKa = 10.88PPQAEE418 pKa = 4.54GEE420 pKa = 4.22NQAFIPEE427 pKa = 4.42DD428 pKa = 3.41TTMVIPVTPAAPVDD442 pKa = 3.71TAPPTEE448 pKa = 4.37VPSEE452 pKa = 3.97YY453 pKa = 10.88AQDD456 pKa = 4.07YY457 pKa = 10.52EE458 pKa = 4.19PGEE461 pKa = 4.15YY462 pKa = 9.95PEE464 pKa = 5.28GDD466 pKa = 3.14EE467 pKa = 4.54SSSEE471 pKa = 3.97EE472 pKa = 4.16TPDD475 pKa = 3.45YY476 pKa = 11.39DD477 pKa = 3.58STFAVDD483 pKa = 3.71QEE485 pKa = 4.27EE486 pKa = 4.89PYY488 pKa = 11.07DD489 pKa = 4.16LEE491 pKa = 4.37EE492 pKa = 5.4SLDD495 pKa = 3.62DD496 pKa = 5.47DD497 pKa = 4.17YY498 pKa = 11.9PEE500 pKa = 5.78EE501 pKa = 4.17ITLDD505 pKa = 3.98DD506 pKa = 4.94APDD509 pKa = 3.93LPVEE513 pKa = 4.43TTDD516 pKa = 3.4VQADD520 pKa = 3.69STALQEE526 pKa = 4.96DD527 pKa = 5.88FYY529 pKa = 11.93APATPEE535 pKa = 4.17EE536 pKa = 4.63TPVAPVAAEE545 pKa = 3.66PAEE548 pKa = 4.34YY549 pKa = 9.93YY550 pKa = 10.82APEE553 pKa = 4.25SVPEE557 pKa = 4.7SISQPYY563 pKa = 8.71VEE565 pKa = 5.19QPQPPYY571 pKa = 9.73ATPEE575 pKa = 4.1SPTIDD580 pKa = 3.86PLTYY584 pKa = 9.39ATPEE588 pKa = 3.87QPAATDD594 pKa = 2.93SWQTPNPYY602 pKa = 9.38QAEE605 pKa = 4.2GLTPSEE611 pKa = 4.07WDD613 pKa = 3.62GTPQPPNYY621 pKa = 9.64EE622 pKa = 4.04QFDD625 pKa = 4.21EE626 pKa = 4.52QPSVQPDD633 pKa = 3.92LQPTDD638 pKa = 3.56QYY640 pKa = 11.69SVPDD644 pKa = 4.89FSANEE649 pKa = 3.82PAYY652 pKa = 9.61ATAAPEE658 pKa = 4.17MPEE661 pKa = 3.95EE662 pKa = 4.11LAVEE666 pKa = 4.27QPEE669 pKa = 4.47PTSEE673 pKa = 4.08TLDD676 pKa = 3.71APTEE680 pKa = 3.84LSEE683 pKa = 4.14PVEE686 pKa = 4.18AAEE689 pKa = 4.05PAEE692 pKa = 4.3PSFSTFQMPQLEE704 pKa = 4.09AVEE707 pKa = 4.3LGKK710 pKa = 10.69DD711 pKa = 3.25EE712 pKa = 4.26VAPVEE717 pKa = 4.33VNDD720 pKa = 5.34AEE722 pKa = 4.41FAPADD727 pKa = 3.63YY728 pKa = 8.4STPTEE733 pKa = 4.22TPKK736 pKa = 11.14APLTDD741 pKa = 3.72VVAPPLPTPGVGLDD755 pKa = 3.53LGQIRR760 pKa = 11.84EE761 pKa = 4.21KK762 pKa = 10.65LAPEE766 pKa = 4.01QADD769 pKa = 3.34SSAEE773 pKa = 4.09SEE775 pKa = 4.29QKK777 pKa = 10.78LADD780 pKa = 3.66VPGVLMTPTRR790 pKa = 11.84SATDD794 pKa = 3.78FPPTEE799 pKa = 4.61SPTEE803 pKa = 4.16SPAAGALTEE812 pKa = 4.29PEE814 pKa = 3.93PTGTVTEE821 pKa = 4.49HH822 pKa = 6.93LSATRR827 pKa = 11.84LEE829 pKa = 4.25EE830 pKa = 4.6LAVQNAASVPEE841 pKa = 4.11TAADD845 pKa = 3.18IPMIDD850 pKa = 3.53TQSEE854 pKa = 4.38NNEE857 pKa = 4.22STKK860 pKa = 10.73GQPTPIFPDD869 pKa = 3.7DD870 pKa = 4.07SNPRR874 pKa = 11.84PMLQGEE880 pKa = 4.36EE881 pKa = 4.17PLAEE885 pKa = 4.07VDD887 pKa = 3.05PDD889 pKa = 3.53GRR891 pKa = 11.84FIPHH895 pKa = 6.66FLAGYY900 pKa = 9.72RR901 pKa = 11.84SNLDD905 pKa = 3.21TFTSVYY911 pKa = 10.72GSLDD915 pKa = 3.42EE916 pKa = 4.54SGSRR920 pKa = 11.84VKK922 pKa = 10.19PASIAKK928 pKa = 10.29LEE930 pKa = 4.23AQQHH934 pKa = 5.49RR935 pKa = 11.84KK936 pKa = 10.16SITTGYY942 pKa = 9.76LVTVATSRR950 pKa = 11.84PLGADD955 pKa = 3.0DD956 pKa = 3.73QVFVRR961 pKa = 11.84LPDD964 pKa = 3.57GRR966 pKa = 11.84EE967 pKa = 3.93VPVTSASSDD976 pKa = 3.28FDD978 pKa = 4.05GVHH981 pKa = 5.48LTIPKK986 pKa = 9.7II987 pKa = 3.71
MM1 pKa = 7.19YY2 pKa = 9.75AQNPALFGKK11 pKa = 10.16QPATLLTVKK20 pKa = 10.25NGKK23 pKa = 8.79DD24 pKa = 2.84RR25 pKa = 11.84HH26 pKa = 5.31MSEE29 pKa = 5.01AAPQLLTSEE38 pKa = 5.64DD39 pKa = 3.29IFSAQFPATKK49 pKa = 10.13FRR51 pKa = 11.84DD52 pKa = 4.43GYY54 pKa = 10.67DD55 pKa = 3.03QNQIDD60 pKa = 4.67DD61 pKa = 4.11YY62 pKa = 10.39LDD64 pKa = 3.29EE65 pKa = 4.38VVRR68 pKa = 11.84VLSYY72 pKa = 11.35YY73 pKa = 10.02EE74 pKa = 4.37ALNASPEE81 pKa = 4.15AEE83 pKa = 3.87VDD85 pKa = 3.38LAYY88 pKa = 9.33ITVRR92 pKa = 11.84GRR94 pKa = 11.84DD95 pKa = 3.45VRR97 pKa = 11.84EE98 pKa = 3.31VDD100 pKa = 3.83FDD102 pKa = 3.77YY103 pKa = 11.3TRR105 pKa = 11.84MRR107 pKa = 11.84VGYY110 pKa = 9.8DD111 pKa = 2.92QDD113 pKa = 4.8AVDD116 pKa = 5.52DD117 pKa = 4.27YY118 pKa = 11.24LDD120 pKa = 3.46QVAATLEE127 pKa = 4.44AYY129 pKa = 7.77EE130 pKa = 4.45KK131 pKa = 10.77LYY133 pKa = 10.72GIPPSDD139 pKa = 3.23QRR141 pKa = 11.84YY142 pKa = 8.28QVLQVDD148 pKa = 5.16GAALEE153 pKa = 4.51AEE155 pKa = 4.48AAEE158 pKa = 4.15QAAASGEE165 pKa = 4.36LPSSSAPNEE174 pKa = 4.15DD175 pKa = 3.61AAVTPIEE182 pKa = 4.49IAAEE186 pKa = 4.26TPPSQPAEE194 pKa = 3.95QTVASAAPLPVTPAATPSEE213 pKa = 4.14DD214 pKa = 3.65LGVSSSILGTPTGSIPVTQPVPFEE238 pKa = 3.84QAVLTNHH245 pKa = 6.78EE246 pKa = 4.79DD247 pKa = 3.58GSPAGHH253 pKa = 6.48EE254 pKa = 4.22FDD256 pKa = 3.9ATSTASSPLFPPLNAAMSPQSQMPEE281 pKa = 3.8TGTNASPVEE290 pKa = 4.56SIPEE294 pKa = 3.76QNFASAALPEE304 pKa = 4.68TEE306 pKa = 4.36TPIAPSVAASGEE318 pKa = 4.16TAPAVPLPEE327 pKa = 5.67PDD329 pKa = 4.6DD330 pKa = 3.62EE331 pKa = 4.5WGEE334 pKa = 4.29VPQYY338 pKa = 10.56PVDD341 pKa = 3.74APEE344 pKa = 4.43PQTPDD349 pKa = 2.48ATAAQTYY356 pKa = 9.51SEE358 pKa = 4.81PGYY361 pKa = 10.82DD362 pKa = 3.2PYY364 pKa = 11.36QPYY367 pKa = 10.39EE368 pKa = 4.03YY369 pKa = 10.27DD370 pKa = 4.8AYY372 pKa = 10.1DD373 pKa = 3.59QPMSTAPAEE382 pKa = 4.29YY383 pKa = 9.62QGSSEE388 pKa = 4.92PYY390 pKa = 9.31PEE392 pKa = 4.96APTEE396 pKa = 3.95AGYY399 pKa = 9.57PSPYY403 pKa = 10.26GAATPEE409 pKa = 4.82GYY411 pKa = 10.09DD412 pKa = 3.55YY413 pKa = 10.88PPQAEE418 pKa = 4.54GEE420 pKa = 4.22NQAFIPEE427 pKa = 4.42DD428 pKa = 3.41TTMVIPVTPAAPVDD442 pKa = 3.71TAPPTEE448 pKa = 4.37VPSEE452 pKa = 3.97YY453 pKa = 10.88AQDD456 pKa = 4.07YY457 pKa = 10.52EE458 pKa = 4.19PGEE461 pKa = 4.15YY462 pKa = 9.95PEE464 pKa = 5.28GDD466 pKa = 3.14EE467 pKa = 4.54SSSEE471 pKa = 3.97EE472 pKa = 4.16TPDD475 pKa = 3.45YY476 pKa = 11.39DD477 pKa = 3.58STFAVDD483 pKa = 3.71QEE485 pKa = 4.27EE486 pKa = 4.89PYY488 pKa = 11.07DD489 pKa = 4.16LEE491 pKa = 4.37EE492 pKa = 5.4SLDD495 pKa = 3.62DD496 pKa = 5.47DD497 pKa = 4.17YY498 pKa = 11.9PEE500 pKa = 5.78EE501 pKa = 4.17ITLDD505 pKa = 3.98DD506 pKa = 4.94APDD509 pKa = 3.93LPVEE513 pKa = 4.43TTDD516 pKa = 3.4VQADD520 pKa = 3.69STALQEE526 pKa = 4.96DD527 pKa = 5.88FYY529 pKa = 11.93APATPEE535 pKa = 4.17EE536 pKa = 4.63TPVAPVAAEE545 pKa = 3.66PAEE548 pKa = 4.34YY549 pKa = 9.93YY550 pKa = 10.82APEE553 pKa = 4.25SVPEE557 pKa = 4.7SISQPYY563 pKa = 8.71VEE565 pKa = 5.19QPQPPYY571 pKa = 9.73ATPEE575 pKa = 4.1SPTIDD580 pKa = 3.86PLTYY584 pKa = 9.39ATPEE588 pKa = 3.87QPAATDD594 pKa = 2.93SWQTPNPYY602 pKa = 9.38QAEE605 pKa = 4.2GLTPSEE611 pKa = 4.07WDD613 pKa = 3.62GTPQPPNYY621 pKa = 9.64EE622 pKa = 4.04QFDD625 pKa = 4.21EE626 pKa = 4.52QPSVQPDD633 pKa = 3.92LQPTDD638 pKa = 3.56QYY640 pKa = 11.69SVPDD644 pKa = 4.89FSANEE649 pKa = 3.82PAYY652 pKa = 9.61ATAAPEE658 pKa = 4.17MPEE661 pKa = 3.95EE662 pKa = 4.11LAVEE666 pKa = 4.27QPEE669 pKa = 4.47PTSEE673 pKa = 4.08TLDD676 pKa = 3.71APTEE680 pKa = 3.84LSEE683 pKa = 4.14PVEE686 pKa = 4.18AAEE689 pKa = 4.05PAEE692 pKa = 4.3PSFSTFQMPQLEE704 pKa = 4.09AVEE707 pKa = 4.3LGKK710 pKa = 10.69DD711 pKa = 3.25EE712 pKa = 4.26VAPVEE717 pKa = 4.33VNDD720 pKa = 5.34AEE722 pKa = 4.41FAPADD727 pKa = 3.63YY728 pKa = 8.4STPTEE733 pKa = 4.22TPKK736 pKa = 11.14APLTDD741 pKa = 3.72VVAPPLPTPGVGLDD755 pKa = 3.53LGQIRR760 pKa = 11.84EE761 pKa = 4.21KK762 pKa = 10.65LAPEE766 pKa = 4.01QADD769 pKa = 3.34SSAEE773 pKa = 4.09SEE775 pKa = 4.29QKK777 pKa = 10.78LADD780 pKa = 3.66VPGVLMTPTRR790 pKa = 11.84SATDD794 pKa = 3.78FPPTEE799 pKa = 4.61SPTEE803 pKa = 4.16SPAAGALTEE812 pKa = 4.29PEE814 pKa = 3.93PTGTVTEE821 pKa = 4.49HH822 pKa = 6.93LSATRR827 pKa = 11.84LEE829 pKa = 4.25EE830 pKa = 4.6LAVQNAASVPEE841 pKa = 4.11TAADD845 pKa = 3.18IPMIDD850 pKa = 3.53TQSEE854 pKa = 4.38NNEE857 pKa = 4.22STKK860 pKa = 10.73GQPTPIFPDD869 pKa = 3.7DD870 pKa = 4.07SNPRR874 pKa = 11.84PMLQGEE880 pKa = 4.36EE881 pKa = 4.17PLAEE885 pKa = 4.07VDD887 pKa = 3.05PDD889 pKa = 3.53GRR891 pKa = 11.84FIPHH895 pKa = 6.66FLAGYY900 pKa = 9.72RR901 pKa = 11.84SNLDD905 pKa = 3.21TFTSVYY911 pKa = 10.72GSLDD915 pKa = 3.42EE916 pKa = 4.54SGSRR920 pKa = 11.84VKK922 pKa = 10.19PASIAKK928 pKa = 10.29LEE930 pKa = 4.23AQQHH934 pKa = 5.49RR935 pKa = 11.84KK936 pKa = 10.16SITTGYY942 pKa = 9.76LVTVATSRR950 pKa = 11.84PLGADD955 pKa = 3.0DD956 pKa = 3.73QVFVRR961 pKa = 11.84LPDD964 pKa = 3.57GRR966 pKa = 11.84EE967 pKa = 3.93VPVTSASSDD976 pKa = 3.28FDD978 pKa = 4.05GVHH981 pKa = 5.48LTIPKK986 pKa = 9.7II987 pKa = 3.71
Molecular weight: 105.62 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|D6ZJH7|D6ZJH7_MOBCV Uncharacterized protein OS=Mobiluncus curtisii (strain ATCC 43063 / DSM 2711 / V125) OX=548479 GN=HMPREF0573_10557 PE=4 SV=1
MM1 pKa = 7.4GSVIKK6 pKa = 10.42KK7 pKa = 8.47RR8 pKa = 11.84RR9 pKa = 11.84KK10 pKa = 9.22RR11 pKa = 11.84MAKK14 pKa = 9.41KK15 pKa = 9.87KK16 pKa = 9.7HH17 pKa = 5.84RR18 pKa = 11.84KK19 pKa = 8.56LLRR22 pKa = 11.84KK23 pKa = 7.78TRR25 pKa = 11.84HH26 pKa = 3.65QRR28 pKa = 11.84RR29 pKa = 11.84NKK31 pKa = 9.78KK32 pKa = 9.85
MM1 pKa = 7.4GSVIKK6 pKa = 10.42KK7 pKa = 8.47RR8 pKa = 11.84RR9 pKa = 11.84KK10 pKa = 9.22RR11 pKa = 11.84MAKK14 pKa = 9.41KK15 pKa = 9.87KK16 pKa = 9.7HH17 pKa = 5.84RR18 pKa = 11.84KK19 pKa = 8.56LLRR22 pKa = 11.84KK23 pKa = 7.78TRR25 pKa = 11.84HH26 pKa = 3.65QRR28 pKa = 11.84RR29 pKa = 11.84NKK31 pKa = 9.78KK32 pKa = 9.85
Molecular weight: 4.08 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
641598 |
32 |
5040 |
337.0 |
36.6 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
11.114 ± 0.072 | 0.861 ± 0.017 |
5.521 ± 0.04 | 5.892 ± 0.056 |
3.329 ± 0.033 | 8.113 ± 0.046 |
1.97 ± 0.026 | 4.995 ± 0.044 |
3.988 ± 0.058 | 9.9 ± 0.073 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.315 ± 0.026 | 3.214 ± 0.038 |
5.246 ± 0.053 | 3.854 ± 0.032 |
5.774 ± 0.056 | 6.034 ± 0.039 |
6.183 ± 0.057 | 7.908 ± 0.045 |
1.404 ± 0.024 | 2.385 ± 0.033 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
