
Hubei dimarhabdovirus virus 3
Taxonomy: Viruses; Riboviria; unclassified Riboviria; unclassified RNA viruses ShiM-2016
Average proteome isoelectric point is 6.19
Get precalculated fractions of proteins
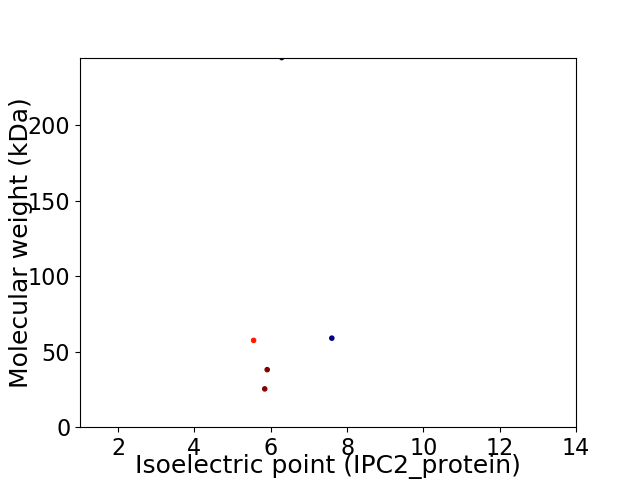
Virtual 2D-PAGE plot for 5 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
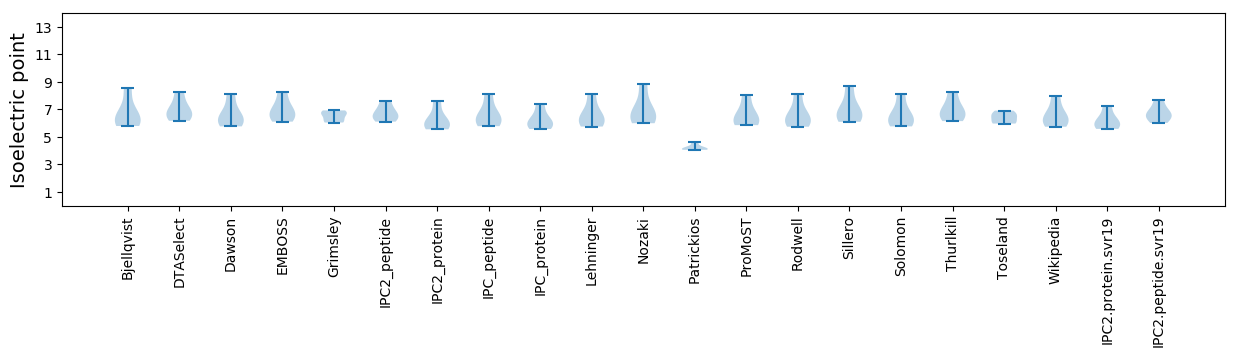
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1L3KMV3|A0A1L3KMV3_9VIRU Uncharacterized protein OS=Hubei dimarhabdovirus virus 3 OX=1922868 PE=4 SV=1
MM1 pKa = 7.7ALNFSDD7 pKa = 4.08EE8 pKa = 4.42RR9 pKa = 11.84FTATSQSMISLYY21 pKa = 10.77EE22 pKa = 3.95SLTNRR27 pKa = 11.84QRR29 pKa = 11.84EE30 pKa = 3.88ILEE33 pKa = 4.74RR34 pKa = 11.84IDD36 pKa = 5.53ANTLSNEE43 pKa = 3.94SLYY46 pKa = 11.34NLDD49 pKa = 3.26QEE51 pKa = 4.69LAAFVPEE58 pKa = 4.15SLPRR62 pKa = 11.84EE63 pKa = 4.22ASPQFPADD71 pKa = 3.35AFRR74 pKa = 11.84AKK76 pKa = 9.06PTVVLYY82 pKa = 10.66SVEE85 pKa = 3.9KK86 pKa = 10.76ADD88 pKa = 3.32VHH90 pKa = 6.61KK91 pKa = 10.91YY92 pKa = 7.66YY93 pKa = 9.17HH94 pKa = 5.88TVMASFRR101 pKa = 11.84EE102 pKa = 4.3KK103 pKa = 10.48LVHH106 pKa = 6.55PKK108 pKa = 9.35MLATFLHH115 pKa = 6.94LSMMTYY121 pKa = 9.69QVEE124 pKa = 4.37LTATLPTAWTSFSIPLTDD142 pKa = 3.87GSLNTSPLHH151 pKa = 6.76LYY153 pKa = 7.82TATAVGTPTLVEE165 pKa = 4.19STVPATEE172 pKa = 3.95KK173 pKa = 10.63AIKK176 pKa = 9.7ALVMVVVGFIRR187 pKa = 11.84YY188 pKa = 9.67SLMIEE193 pKa = 3.84NLGDD197 pKa = 3.29YY198 pKa = 10.21RR199 pKa = 11.84RR200 pKa = 11.84NFNTRR205 pKa = 11.84LTEE208 pKa = 3.99MVKK211 pKa = 10.67SIDD214 pKa = 3.49PDD216 pKa = 4.0FPADD220 pKa = 3.33VSSYY224 pKa = 10.89SGIRR228 pKa = 11.84GMNNFFPDD236 pKa = 3.8LGLVAAALDD245 pKa = 4.29MYY247 pKa = 8.5TVRR250 pKa = 11.84VPQSEE255 pKa = 3.7WRR257 pKa = 11.84IFNMGTLVLRR267 pKa = 11.84NMSAAALVAVTYY279 pKa = 9.59MRR281 pKa = 11.84EE282 pKa = 3.92IMNSTEE288 pKa = 4.02SNHH291 pKa = 6.21YY292 pKa = 7.18YY293 pKa = 10.52HH294 pKa = 6.68SWIWVQSAADD304 pKa = 4.18EE305 pKa = 4.28YY306 pKa = 11.08AKK308 pKa = 10.19MMRR311 pKa = 11.84SGQEE315 pKa = 3.38INNLYY320 pKa = 10.65SYY322 pKa = 11.29APYY325 pKa = 11.36LMTLGKK331 pKa = 10.01AVKK334 pKa = 9.57SPYY337 pKa = 9.98SATVNSNLFTFIHH350 pKa = 6.38VIGASLGLPRR360 pKa = 11.84SINSRR365 pKa = 11.84QLDD368 pKa = 3.97HH369 pKa = 7.26INLNEE374 pKa = 4.53LIINAQFYY382 pKa = 10.66VFAAQRR388 pKa = 11.84APRR391 pKa = 11.84FNMHH395 pKa = 5.47VVNDD399 pKa = 4.05EE400 pKa = 4.13IIRR403 pKa = 11.84ALKK406 pKa = 9.67EE407 pKa = 3.3IHH409 pKa = 6.53AGNRR413 pKa = 11.84AAVGQLAEE421 pKa = 3.77IRR423 pKa = 11.84GRR425 pKa = 11.84LVEE428 pKa = 4.51NDD430 pKa = 3.72DD431 pKa = 3.99EE432 pKa = 4.45EE433 pKa = 4.74MEE435 pKa = 4.85IEE437 pKa = 4.74DD438 pKa = 4.04SASVAGTNDD447 pKa = 2.87LTQFQTAVDD456 pKa = 3.71HH457 pKa = 5.88EE458 pKa = 4.69AYY460 pKa = 9.21PRR462 pKa = 11.84SSSGVEE468 pKa = 3.36WSSWYY473 pKa = 10.0FRR475 pKa = 11.84NKK477 pKa = 9.27VVPEE481 pKa = 3.93EE482 pKa = 4.08VKK484 pKa = 10.83EE485 pKa = 4.01NCQKK489 pKa = 10.0VWRR492 pKa = 11.84QFVTPRR498 pKa = 11.84NNSLGSVLKK507 pKa = 10.76NLSS510 pKa = 3.21
MM1 pKa = 7.7ALNFSDD7 pKa = 4.08EE8 pKa = 4.42RR9 pKa = 11.84FTATSQSMISLYY21 pKa = 10.77EE22 pKa = 3.95SLTNRR27 pKa = 11.84QRR29 pKa = 11.84EE30 pKa = 3.88ILEE33 pKa = 4.74RR34 pKa = 11.84IDD36 pKa = 5.53ANTLSNEE43 pKa = 3.94SLYY46 pKa = 11.34NLDD49 pKa = 3.26QEE51 pKa = 4.69LAAFVPEE58 pKa = 4.15SLPRR62 pKa = 11.84EE63 pKa = 4.22ASPQFPADD71 pKa = 3.35AFRR74 pKa = 11.84AKK76 pKa = 9.06PTVVLYY82 pKa = 10.66SVEE85 pKa = 3.9KK86 pKa = 10.76ADD88 pKa = 3.32VHH90 pKa = 6.61KK91 pKa = 10.91YY92 pKa = 7.66YY93 pKa = 9.17HH94 pKa = 5.88TVMASFRR101 pKa = 11.84EE102 pKa = 4.3KK103 pKa = 10.48LVHH106 pKa = 6.55PKK108 pKa = 9.35MLATFLHH115 pKa = 6.94LSMMTYY121 pKa = 9.69QVEE124 pKa = 4.37LTATLPTAWTSFSIPLTDD142 pKa = 3.87GSLNTSPLHH151 pKa = 6.76LYY153 pKa = 7.82TATAVGTPTLVEE165 pKa = 4.19STVPATEE172 pKa = 3.95KK173 pKa = 10.63AIKK176 pKa = 9.7ALVMVVVGFIRR187 pKa = 11.84YY188 pKa = 9.67SLMIEE193 pKa = 3.84NLGDD197 pKa = 3.29YY198 pKa = 10.21RR199 pKa = 11.84RR200 pKa = 11.84NFNTRR205 pKa = 11.84LTEE208 pKa = 3.99MVKK211 pKa = 10.67SIDD214 pKa = 3.49PDD216 pKa = 4.0FPADD220 pKa = 3.33VSSYY224 pKa = 10.89SGIRR228 pKa = 11.84GMNNFFPDD236 pKa = 3.8LGLVAAALDD245 pKa = 4.29MYY247 pKa = 8.5TVRR250 pKa = 11.84VPQSEE255 pKa = 3.7WRR257 pKa = 11.84IFNMGTLVLRR267 pKa = 11.84NMSAAALVAVTYY279 pKa = 9.59MRR281 pKa = 11.84EE282 pKa = 3.92IMNSTEE288 pKa = 4.02SNHH291 pKa = 6.21YY292 pKa = 7.18YY293 pKa = 10.52HH294 pKa = 6.68SWIWVQSAADD304 pKa = 4.18EE305 pKa = 4.28YY306 pKa = 11.08AKK308 pKa = 10.19MMRR311 pKa = 11.84SGQEE315 pKa = 3.38INNLYY320 pKa = 10.65SYY322 pKa = 11.29APYY325 pKa = 11.36LMTLGKK331 pKa = 10.01AVKK334 pKa = 9.57SPYY337 pKa = 9.98SATVNSNLFTFIHH350 pKa = 6.38VIGASLGLPRR360 pKa = 11.84SINSRR365 pKa = 11.84QLDD368 pKa = 3.97HH369 pKa = 7.26INLNEE374 pKa = 4.53LIINAQFYY382 pKa = 10.66VFAAQRR388 pKa = 11.84APRR391 pKa = 11.84FNMHH395 pKa = 5.47VVNDD399 pKa = 4.05EE400 pKa = 4.13IIRR403 pKa = 11.84ALKK406 pKa = 9.67EE407 pKa = 3.3IHH409 pKa = 6.53AGNRR413 pKa = 11.84AAVGQLAEE421 pKa = 3.77IRR423 pKa = 11.84GRR425 pKa = 11.84LVEE428 pKa = 4.51NDD430 pKa = 3.72DD431 pKa = 3.99EE432 pKa = 4.45EE433 pKa = 4.74MEE435 pKa = 4.85IEE437 pKa = 4.74DD438 pKa = 4.04SASVAGTNDD447 pKa = 2.87LTQFQTAVDD456 pKa = 3.71HH457 pKa = 5.88EE458 pKa = 4.69AYY460 pKa = 9.21PRR462 pKa = 11.84SSSGVEE468 pKa = 3.36WSSWYY473 pKa = 10.0FRR475 pKa = 11.84NKK477 pKa = 9.27VVPEE481 pKa = 3.93EE482 pKa = 4.08VKK484 pKa = 10.83EE485 pKa = 4.01NCQKK489 pKa = 10.0VWRR492 pKa = 11.84QFVTPRR498 pKa = 11.84NNSLGSVLKK507 pKa = 10.76NLSS510 pKa = 3.21
Molecular weight: 57.55 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1L3KMX0|A0A1L3KMX0_9VIRU Uncharacterized protein OS=Hubei dimarhabdovirus virus 3 OX=1922868 PE=4 SV=1
MM1 pKa = 7.68KK2 pKa = 9.15KK3 pKa = 8.68TNKK6 pKa = 6.8NHH8 pKa = 5.68KK9 pKa = 8.56MKK11 pKa = 10.33RR12 pKa = 11.84LNRR15 pKa = 11.84IRR17 pKa = 11.84SLLILLIIGEE27 pKa = 4.4RR28 pKa = 11.84TSSILLPSYY37 pKa = 10.09EE38 pKa = 4.0PKK40 pKa = 9.5QWEE43 pKa = 4.26RR44 pKa = 11.84VNPIDD49 pKa = 4.67LSCPRR54 pKa = 11.84IEE56 pKa = 4.31SVKK59 pKa = 10.27TEE61 pKa = 4.08VVDD64 pKa = 4.78ISPSQCSRR72 pKa = 11.84VSRR75 pKa = 11.84ALTRR79 pKa = 11.84QEE81 pKa = 3.85SGFWCIGVVMITKK94 pKa = 8.85CTEE97 pKa = 3.7SCFGGTDD104 pKa = 3.37VVRR107 pKa = 11.84VSRR110 pKa = 11.84DD111 pKa = 3.15NPVSYY116 pKa = 9.67TEE118 pKa = 5.13CSDD121 pKa = 3.88ALQDD125 pKa = 3.82FLKK128 pKa = 10.88GQLSFPAFPPAQCSWCSSNSEE149 pKa = 3.98EE150 pKa = 4.54AKK152 pKa = 10.67SILIVKK158 pKa = 9.81EE159 pKa = 4.23DD160 pKa = 3.46IPYY163 pKa = 10.6EE164 pKa = 4.15PLTNLYY170 pKa = 9.19PLGGIDD176 pKa = 3.98PTSCDD181 pKa = 3.04NHH183 pKa = 6.55HH184 pKa = 7.06CLTNSRR190 pKa = 11.84SRR192 pKa = 11.84MWMRR196 pKa = 11.84WTTNSEE202 pKa = 4.23SCFLQPEE209 pKa = 4.3NFLLLRR215 pKa = 11.84NRR217 pKa = 11.84TGSYY221 pKa = 10.51LRR223 pKa = 11.84DD224 pKa = 3.77SVHH227 pKa = 5.69RR228 pKa = 11.84TYY230 pKa = 11.55SLLSACLTDD239 pKa = 3.68YY240 pKa = 10.85CHH242 pKa = 7.39RR243 pKa = 11.84PTLYY247 pKa = 10.77LSGRR251 pKa = 11.84EE252 pKa = 3.96LLSCEE257 pKa = 4.74SVLLDD262 pKa = 3.32QLKK265 pKa = 10.63LKK267 pKa = 10.28KK268 pKa = 10.04CPKK271 pKa = 8.48QIRR274 pKa = 11.84YY275 pKa = 9.45EE276 pKa = 3.96IDD278 pKa = 3.24PANQDD283 pKa = 3.41VLAEE287 pKa = 4.02DD288 pKa = 3.68QFIQRR293 pKa = 11.84EE294 pKa = 4.21LVQVKK299 pKa = 10.45CEE301 pKa = 3.98DD302 pKa = 3.17SRR304 pKa = 11.84EE305 pKa = 3.88RR306 pKa = 11.84VIKK309 pKa = 10.31GGSIAPSDD317 pKa = 4.25LYY319 pKa = 11.69LMSKK323 pKa = 9.91KK324 pKa = 10.69GPGEE328 pKa = 4.11GYY330 pKa = 10.91VYY332 pKa = 10.53FVDD335 pKa = 3.41NAGVLKK341 pKa = 10.73RR342 pKa = 11.84GLTNYY347 pKa = 10.1RR348 pKa = 11.84EE349 pKa = 4.14VVAAEE354 pKa = 4.09SMVNLTSSLSSSEE367 pKa = 4.21TIKK370 pKa = 10.39WSLWTKK376 pKa = 7.86LTPPGLFMGPNGIVWDD392 pKa = 4.08EE393 pKa = 3.93TSEE396 pKa = 4.12RR397 pKa = 11.84LSFPSLFYY405 pKa = 11.22DD406 pKa = 3.68RR407 pKa = 11.84VLNLDD412 pKa = 3.48VKK414 pKa = 10.05LANMTQTLSDD424 pKa = 3.8GADD427 pKa = 3.47HH428 pKa = 5.79QTHH431 pKa = 7.15PGLGSLLKK439 pKa = 11.07NMILQHH445 pKa = 6.07DD446 pKa = 4.07HH447 pKa = 6.67SKK449 pKa = 11.34NILDD453 pKa = 3.92VVATAGGDD461 pKa = 3.74LVTSTILTVTKK472 pKa = 10.24CVLYY476 pKa = 10.4ILGLIIFIQVTACVLRR492 pKa = 11.84KK493 pKa = 9.57VLCSPKK499 pKa = 10.04KK500 pKa = 9.91GKK502 pKa = 8.06TPQGYY507 pKa = 9.95DD508 pKa = 2.93LVRR511 pKa = 11.84YY512 pKa = 9.99VNGPQAAVSISKK524 pKa = 10.38ASKK527 pKa = 9.91
MM1 pKa = 7.68KK2 pKa = 9.15KK3 pKa = 8.68TNKK6 pKa = 6.8NHH8 pKa = 5.68KK9 pKa = 8.56MKK11 pKa = 10.33RR12 pKa = 11.84LNRR15 pKa = 11.84IRR17 pKa = 11.84SLLILLIIGEE27 pKa = 4.4RR28 pKa = 11.84TSSILLPSYY37 pKa = 10.09EE38 pKa = 4.0PKK40 pKa = 9.5QWEE43 pKa = 4.26RR44 pKa = 11.84VNPIDD49 pKa = 4.67LSCPRR54 pKa = 11.84IEE56 pKa = 4.31SVKK59 pKa = 10.27TEE61 pKa = 4.08VVDD64 pKa = 4.78ISPSQCSRR72 pKa = 11.84VSRR75 pKa = 11.84ALTRR79 pKa = 11.84QEE81 pKa = 3.85SGFWCIGVVMITKK94 pKa = 8.85CTEE97 pKa = 3.7SCFGGTDD104 pKa = 3.37VVRR107 pKa = 11.84VSRR110 pKa = 11.84DD111 pKa = 3.15NPVSYY116 pKa = 9.67TEE118 pKa = 5.13CSDD121 pKa = 3.88ALQDD125 pKa = 3.82FLKK128 pKa = 10.88GQLSFPAFPPAQCSWCSSNSEE149 pKa = 3.98EE150 pKa = 4.54AKK152 pKa = 10.67SILIVKK158 pKa = 9.81EE159 pKa = 4.23DD160 pKa = 3.46IPYY163 pKa = 10.6EE164 pKa = 4.15PLTNLYY170 pKa = 9.19PLGGIDD176 pKa = 3.98PTSCDD181 pKa = 3.04NHH183 pKa = 6.55HH184 pKa = 7.06CLTNSRR190 pKa = 11.84SRR192 pKa = 11.84MWMRR196 pKa = 11.84WTTNSEE202 pKa = 4.23SCFLQPEE209 pKa = 4.3NFLLLRR215 pKa = 11.84NRR217 pKa = 11.84TGSYY221 pKa = 10.51LRR223 pKa = 11.84DD224 pKa = 3.77SVHH227 pKa = 5.69RR228 pKa = 11.84TYY230 pKa = 11.55SLLSACLTDD239 pKa = 3.68YY240 pKa = 10.85CHH242 pKa = 7.39RR243 pKa = 11.84PTLYY247 pKa = 10.77LSGRR251 pKa = 11.84EE252 pKa = 3.96LLSCEE257 pKa = 4.74SVLLDD262 pKa = 3.32QLKK265 pKa = 10.63LKK267 pKa = 10.28KK268 pKa = 10.04CPKK271 pKa = 8.48QIRR274 pKa = 11.84YY275 pKa = 9.45EE276 pKa = 3.96IDD278 pKa = 3.24PANQDD283 pKa = 3.41VLAEE287 pKa = 4.02DD288 pKa = 3.68QFIQRR293 pKa = 11.84EE294 pKa = 4.21LVQVKK299 pKa = 10.45CEE301 pKa = 3.98DD302 pKa = 3.17SRR304 pKa = 11.84EE305 pKa = 3.88RR306 pKa = 11.84VIKK309 pKa = 10.31GGSIAPSDD317 pKa = 4.25LYY319 pKa = 11.69LMSKK323 pKa = 9.91KK324 pKa = 10.69GPGEE328 pKa = 4.11GYY330 pKa = 10.91VYY332 pKa = 10.53FVDD335 pKa = 3.41NAGVLKK341 pKa = 10.73RR342 pKa = 11.84GLTNYY347 pKa = 10.1RR348 pKa = 11.84EE349 pKa = 4.14VVAAEE354 pKa = 4.09SMVNLTSSLSSSEE367 pKa = 4.21TIKK370 pKa = 10.39WSLWTKK376 pKa = 7.86LTPPGLFMGPNGIVWDD392 pKa = 4.08EE393 pKa = 3.93TSEE396 pKa = 4.12RR397 pKa = 11.84LSFPSLFYY405 pKa = 11.22DD406 pKa = 3.68RR407 pKa = 11.84VLNLDD412 pKa = 3.48VKK414 pKa = 10.05LANMTQTLSDD424 pKa = 3.8GADD427 pKa = 3.47HH428 pKa = 5.79QTHH431 pKa = 7.15PGLGSLLKK439 pKa = 11.07NMILQHH445 pKa = 6.07DD446 pKa = 4.07HH447 pKa = 6.67SKK449 pKa = 11.34NILDD453 pKa = 3.92VVATAGGDD461 pKa = 3.74LVTSTILTVTKK472 pKa = 10.24CVLYY476 pKa = 10.4ILGLIIFIQVTACVLRR492 pKa = 11.84KK493 pKa = 9.57VLCSPKK499 pKa = 10.04KK500 pKa = 9.91GKK502 pKa = 8.06TPQGYY507 pKa = 9.95DD508 pKa = 2.93LVRR511 pKa = 11.84YY512 pKa = 9.99VNGPQAAVSISKK524 pKa = 10.38ASKK527 pKa = 9.91
Molecular weight: 59.03 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
3740 |
224 |
2134 |
748.0 |
85.03 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
5.856 ± 0.985 | 1.845 ± 0.459 |
5.241 ± 0.289 | 6.337 ± 0.329 |
4.358 ± 0.445 | 4.947 ± 0.405 |
2.888 ± 0.417 | 6.123 ± 0.685 |
5.134 ± 0.563 | 9.893 ± 0.651 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.807 ± 0.294 | 4.786 ± 0.366 |
5.08 ± 0.667 | 3.583 ± 0.295 |
5.963 ± 0.208 | 8.85 ± 0.539 |
5.668 ± 0.222 | 5.668 ± 0.643 |
1.364 ± 0.16 | 3.61 ± 0.302 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
