
Paracoccus sp. BM15
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Alphaproteobacteria; Rhodobacterales; Rhodobacteraceae; Paracoccus; unclassified Paracoccus
Average proteome isoelectric point is 6.11
Get precalculated fractions of proteins
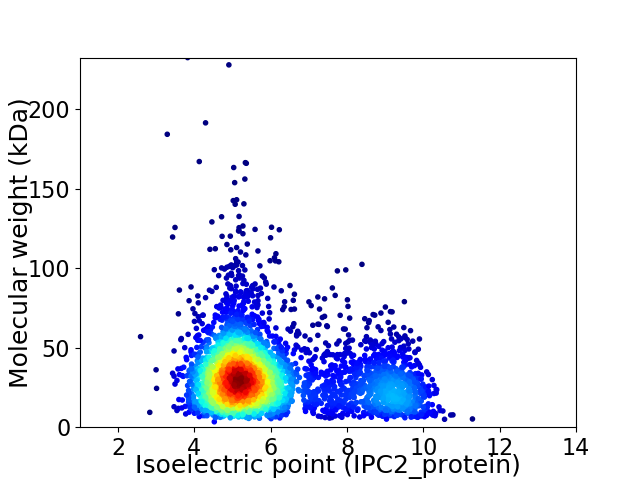
Virtual 2D-PAGE plot for 3740 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A2K9EEP5|A0A2K9EEP5_9RHOB 3-mercaptopyruvate sulfurtransferase OS=Paracoccus sp. BM15 OX=1529068 GN=CUV01_08485 PE=4 SV=1
MM1 pKa = 7.44KK2 pKa = 10.19KK3 pKa = 10.41VLFATTALVMTAGVAAAEE21 pKa = 4.39VSLSGDD27 pKa = 2.74GRR29 pKa = 11.84MGMVYY34 pKa = 10.52DD35 pKa = 4.8GDD37 pKa = 4.47DD38 pKa = 3.41LQFTSRR44 pKa = 11.84ARR46 pKa = 11.84VKK48 pKa = 9.79FTLTGEE54 pKa = 4.24SDD56 pKa = 3.24SGLSFGGEE64 pKa = 3.52FRR66 pKa = 11.84VDD68 pKa = 4.06HH69 pKa = 6.7EE70 pKa = 5.26DD71 pKa = 3.41EE72 pKa = 5.52DD73 pKa = 4.69GSSASRR79 pKa = 11.84GGAGHH84 pKa = 6.52VYY86 pKa = 10.41ISGAYY91 pKa = 9.13GKK93 pKa = 10.77LSMGDD98 pKa = 2.88IDD100 pKa = 4.85SASEE104 pKa = 3.88KK105 pKa = 11.15ANGDD109 pKa = 3.42LHH111 pKa = 7.27GVGLTGLGDD120 pKa = 3.41INEE123 pKa = 4.33FVYY126 pKa = 9.41LTSDD130 pKa = 4.42FEE132 pKa = 5.75SNDD135 pKa = 3.44NPGALYY141 pKa = 10.2EE142 pKa = 4.27YY143 pKa = 7.67TTGAFTAYY151 pKa = 10.53ASMMDD156 pKa = 3.63AGDD159 pKa = 4.62DD160 pKa = 3.64YY161 pKa = 11.89GFDD164 pKa = 4.05GDD166 pKa = 5.63NDD168 pKa = 4.66TVWSIGGKK176 pKa = 9.46YY177 pKa = 9.37AASNYY182 pKa = 10.29AFGLGYY188 pKa = 10.34EE189 pKa = 4.3AVDD192 pKa = 3.81VDD194 pKa = 5.75GFDD197 pKa = 3.93LNQWTISGEE206 pKa = 4.33GTFGQATVKK215 pKa = 10.62AIYY218 pKa = 10.5SDD220 pKa = 3.63MDD222 pKa = 3.59NDD224 pKa = 4.81GVFDD228 pKa = 4.56DD229 pKa = 4.1VKK231 pKa = 11.02QYY233 pKa = 11.11GLSVMYY239 pKa = 10.34DD240 pKa = 3.25AGATDD245 pKa = 3.53VSAFYY250 pKa = 10.95KK251 pKa = 10.13KK252 pKa = 10.37DD253 pKa = 3.29EE254 pKa = 4.08YY255 pKa = 11.66SFAGVDD261 pKa = 3.32TGDD264 pKa = 3.47ADD266 pKa = 3.46TWGIGAAYY274 pKa = 10.02DD275 pKa = 3.78LGGGATLKK283 pKa = 11.01GGIVDD288 pKa = 3.52SDD290 pKa = 4.1YY291 pKa = 11.64NADD294 pKa = 3.9TIADD298 pKa = 3.94FGVAFTFF305 pKa = 4.36
MM1 pKa = 7.44KK2 pKa = 10.19KK3 pKa = 10.41VLFATTALVMTAGVAAAEE21 pKa = 4.39VSLSGDD27 pKa = 2.74GRR29 pKa = 11.84MGMVYY34 pKa = 10.52DD35 pKa = 4.8GDD37 pKa = 4.47DD38 pKa = 3.41LQFTSRR44 pKa = 11.84ARR46 pKa = 11.84VKK48 pKa = 9.79FTLTGEE54 pKa = 4.24SDD56 pKa = 3.24SGLSFGGEE64 pKa = 3.52FRR66 pKa = 11.84VDD68 pKa = 4.06HH69 pKa = 6.7EE70 pKa = 5.26DD71 pKa = 3.41EE72 pKa = 5.52DD73 pKa = 4.69GSSASRR79 pKa = 11.84GGAGHH84 pKa = 6.52VYY86 pKa = 10.41ISGAYY91 pKa = 9.13GKK93 pKa = 10.77LSMGDD98 pKa = 2.88IDD100 pKa = 4.85SASEE104 pKa = 3.88KK105 pKa = 11.15ANGDD109 pKa = 3.42LHH111 pKa = 7.27GVGLTGLGDD120 pKa = 3.41INEE123 pKa = 4.33FVYY126 pKa = 9.41LTSDD130 pKa = 4.42FEE132 pKa = 5.75SNDD135 pKa = 3.44NPGALYY141 pKa = 10.2EE142 pKa = 4.27YY143 pKa = 7.67TTGAFTAYY151 pKa = 10.53ASMMDD156 pKa = 3.63AGDD159 pKa = 4.62DD160 pKa = 3.64YY161 pKa = 11.89GFDD164 pKa = 4.05GDD166 pKa = 5.63NDD168 pKa = 4.66TVWSIGGKK176 pKa = 9.46YY177 pKa = 9.37AASNYY182 pKa = 10.29AFGLGYY188 pKa = 10.34EE189 pKa = 4.3AVDD192 pKa = 3.81VDD194 pKa = 5.75GFDD197 pKa = 3.93LNQWTISGEE206 pKa = 4.33GTFGQATVKK215 pKa = 10.62AIYY218 pKa = 10.5SDD220 pKa = 3.63MDD222 pKa = 3.59NDD224 pKa = 4.81GVFDD228 pKa = 4.56DD229 pKa = 4.1VKK231 pKa = 11.02QYY233 pKa = 11.11GLSVMYY239 pKa = 10.34DD240 pKa = 3.25AGATDD245 pKa = 3.53VSAFYY250 pKa = 10.95KK251 pKa = 10.13KK252 pKa = 10.37DD253 pKa = 3.29EE254 pKa = 4.08YY255 pKa = 11.66SFAGVDD261 pKa = 3.32TGDD264 pKa = 3.47ADD266 pKa = 3.46TWGIGAAYY274 pKa = 10.02DD275 pKa = 3.78LGGGATLKK283 pKa = 11.01GGIVDD288 pKa = 3.52SDD290 pKa = 4.1YY291 pKa = 11.64NADD294 pKa = 3.9TIADD298 pKa = 3.94FGVAFTFF305 pKa = 4.36
Molecular weight: 31.96 kDa
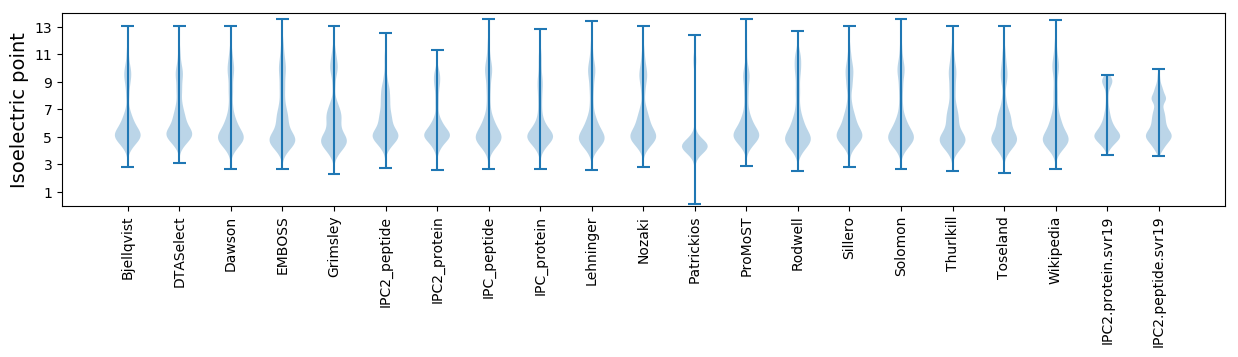
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A2K9ECQ1|A0A2K9ECQ1_9RHOB GNAT family N-acetyltransferase OS=Paracoccus sp. BM15 OX=1529068 GN=CUV01_04280 PE=4 SV=1
MM1 pKa = 7.45KK2 pKa = 9.61RR3 pKa = 11.84TFQPSNLVRR12 pKa = 11.84ARR14 pKa = 11.84RR15 pKa = 11.84HH16 pKa = 4.37GFRR19 pKa = 11.84ARR21 pKa = 11.84MATKK25 pKa = 10.41AGRR28 pKa = 11.84LVLNRR33 pKa = 11.84RR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 9.7GRR39 pKa = 11.84KK40 pKa = 8.85RR41 pKa = 11.84LSAA44 pKa = 3.96
MM1 pKa = 7.45KK2 pKa = 9.61RR3 pKa = 11.84TFQPSNLVRR12 pKa = 11.84ARR14 pKa = 11.84RR15 pKa = 11.84HH16 pKa = 4.37GFRR19 pKa = 11.84ARR21 pKa = 11.84MATKK25 pKa = 10.41AGRR28 pKa = 11.84LVLNRR33 pKa = 11.84RR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 9.7GRR39 pKa = 11.84KK40 pKa = 8.85RR41 pKa = 11.84LSAA44 pKa = 3.96
Molecular weight: 5.18 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1166477 |
30 |
2290 |
311.9 |
33.82 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
12.552 ± 0.062 | 0.823 ± 0.011 |
6.454 ± 0.039 | 5.547 ± 0.037 |
3.569 ± 0.024 | 8.87 ± 0.041 |
2.0 ± 0.019 | 5.326 ± 0.029 |
2.779 ± 0.035 | 9.968 ± 0.049 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.876 ± 0.022 | 2.59 ± 0.025 |
5.228 ± 0.031 | 3.567 ± 0.024 |
6.956 ± 0.039 | 5.103 ± 0.024 |
5.299 ± 0.029 | 6.901 ± 0.033 |
1.476 ± 0.018 | 2.115 ± 0.018 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
