
Methylovorus glucosetrophus (strain SIP3-4)
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Betaproteobacteria; Nitrosomonadales; Methylophilaceae; Methylovorus; Methylovorus glucosotrophus
Average proteome isoelectric point is 6.5
Get precalculated fractions of proteins
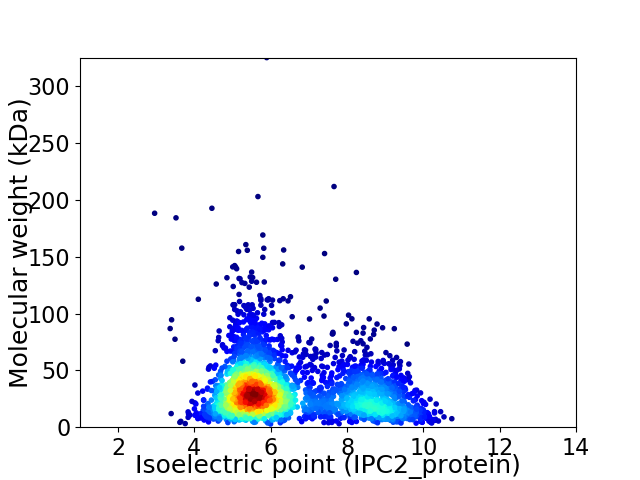
Virtual 2D-PAGE plot for 2907 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|C6XBQ3|C6XBQ3_METGS Acyl carrier protein-related protein OS=Methylovorus glucosetrophus (strain SIP3-4) OX=582744 GN=Msip34_2786 PE=4 SV=1
MM1 pKa = 7.52AKK3 pKa = 8.77STSFDD8 pKa = 3.59DD9 pKa = 3.76YY10 pKa = 10.67VTYY13 pKa = 10.89LIDD16 pKa = 3.62TEE18 pKa = 4.51EE19 pKa = 4.65NDD21 pKa = 3.61TASASLGKK29 pKa = 7.45TTITVSLAGLTDD41 pKa = 3.55NSQQEE46 pKa = 3.92AAARR50 pKa = 11.84HH51 pKa = 5.61ALEE54 pKa = 4.21AWEE57 pKa = 4.91DD58 pKa = 3.63ATGLTFTIVTSGSADD73 pKa = 2.97IYY75 pKa = 10.73IGNDD79 pKa = 3.2DD80 pKa = 4.29ALGEE84 pKa = 3.94GAYY87 pKa = 9.99AYY89 pKa = 9.74EE90 pKa = 4.53EE91 pKa = 3.81NDD93 pKa = 2.8ARR95 pKa = 11.84YY96 pKa = 10.32INVSEE101 pKa = 3.81NWMDD105 pKa = 3.99GFPADD110 pKa = 4.25EE111 pKa = 4.03QWSVGSYY118 pKa = 9.81GVQTFIHH125 pKa = 6.8EE126 pKa = 4.31IGHH129 pKa = 5.17TLGLGHH135 pKa = 7.29GGPYY139 pKa = 9.97NGNGSYY145 pKa = 11.03AKK147 pKa = 10.36DD148 pKa = 3.47AIFALDD154 pKa = 3.55TNQYY158 pKa = 11.1SIMSYY163 pKa = 10.65FDD165 pKa = 2.99QSNYY169 pKa = 10.35GGASNLFLLSPMMADD184 pKa = 3.26IAAIRR189 pKa = 11.84LLYY192 pKa = 10.85GNLATNTGDD201 pKa = 3.2TLYY204 pKa = 11.45GLGEE208 pKa = 4.3TVLGGTSDD216 pKa = 3.24MGSYY220 pKa = 10.36AGYY223 pKa = 10.83ARR225 pKa = 11.84TISDD229 pKa = 3.42SDD231 pKa = 3.67GTDD234 pKa = 2.71TMDD237 pKa = 4.05FSNVSRR243 pKa = 11.84GSVIDD248 pKa = 3.4MRR250 pKa = 11.84AGGFSSINGYY260 pKa = 9.75KK261 pKa = 10.33KK262 pKa = 10.56NFAIAQDD269 pKa = 3.76TTIEE273 pKa = 4.17NLNATLGRR281 pKa = 11.84DD282 pKa = 3.34TVTGNDD288 pKa = 4.11ADD290 pKa = 3.85NLIYY294 pKa = 10.65GLSGNDD300 pKa = 4.09KK301 pKa = 10.14ISSGAGNDD309 pKa = 3.72TLDD312 pKa = 4.03GGLGGDD318 pKa = 4.07TLTGGLGDD326 pKa = 3.86DD327 pKa = 3.7VYY329 pKa = 11.41YY330 pKa = 10.45INSSKK335 pKa = 11.13DD336 pKa = 3.59VISEE340 pKa = 4.19KK341 pKa = 10.88ADD343 pKa = 3.24GGTDD347 pKa = 3.25VVYY350 pKa = 11.02ASVSYY355 pKa = 10.59TMASYY360 pKa = 10.96LEE362 pKa = 4.21EE363 pKa = 4.1VVLQGSASTKK373 pKa = 10.62LKK375 pKa = 9.79ATGNAMANQITGDD388 pKa = 3.7DD389 pKa = 3.9GANVLNGGAGNDD401 pKa = 3.72TLAGGLGRR409 pKa = 11.84DD410 pKa = 3.7TLAGGTGNDD419 pKa = 3.83SLDD422 pKa = 3.95GGDD425 pKa = 5.51ANDD428 pKa = 4.7KK429 pKa = 10.92LDD431 pKa = 4.31GGAGDD436 pKa = 3.94DD437 pKa = 4.16TLVGGAGNDD446 pKa = 3.81LLKK449 pKa = 11.08GGAGSDD455 pKa = 2.99IFIIEE460 pKa = 5.36DD461 pKa = 3.73GTDD464 pKa = 2.94HH465 pKa = 8.02DD466 pKa = 5.18IISDD470 pKa = 3.97FAYY473 pKa = 10.5SSKK476 pKa = 9.46TSKK479 pKa = 11.08GITTIVEE486 pKa = 3.96DD487 pKa = 3.36KK488 pKa = 11.12LYY490 pKa = 9.45ITGHH494 pKa = 7.11DD495 pKa = 3.88YY496 pKa = 11.43DD497 pKa = 4.33QDD499 pKa = 3.97TLFDD503 pKa = 4.55GDD505 pKa = 4.54DD506 pKa = 3.79YY507 pKa = 11.98SVGLAADD514 pKa = 3.87DD515 pKa = 4.17SGLLFSFYY523 pKa = 10.89SGGSLLLKK531 pKa = 10.84GIGEE535 pKa = 4.35SMLDD539 pKa = 3.38KK540 pKa = 10.87FDD542 pKa = 3.53IAALHH547 pKa = 6.0TDD549 pKa = 3.76EE550 pKa = 6.1NGDD553 pKa = 3.86SYY555 pKa = 11.75FSLAA559 pKa = 4.01
MM1 pKa = 7.52AKK3 pKa = 8.77STSFDD8 pKa = 3.59DD9 pKa = 3.76YY10 pKa = 10.67VTYY13 pKa = 10.89LIDD16 pKa = 3.62TEE18 pKa = 4.51EE19 pKa = 4.65NDD21 pKa = 3.61TASASLGKK29 pKa = 7.45TTITVSLAGLTDD41 pKa = 3.55NSQQEE46 pKa = 3.92AAARR50 pKa = 11.84HH51 pKa = 5.61ALEE54 pKa = 4.21AWEE57 pKa = 4.91DD58 pKa = 3.63ATGLTFTIVTSGSADD73 pKa = 2.97IYY75 pKa = 10.73IGNDD79 pKa = 3.2DD80 pKa = 4.29ALGEE84 pKa = 3.94GAYY87 pKa = 9.99AYY89 pKa = 9.74EE90 pKa = 4.53EE91 pKa = 3.81NDD93 pKa = 2.8ARR95 pKa = 11.84YY96 pKa = 10.32INVSEE101 pKa = 3.81NWMDD105 pKa = 3.99GFPADD110 pKa = 4.25EE111 pKa = 4.03QWSVGSYY118 pKa = 9.81GVQTFIHH125 pKa = 6.8EE126 pKa = 4.31IGHH129 pKa = 5.17TLGLGHH135 pKa = 7.29GGPYY139 pKa = 9.97NGNGSYY145 pKa = 11.03AKK147 pKa = 10.36DD148 pKa = 3.47AIFALDD154 pKa = 3.55TNQYY158 pKa = 11.1SIMSYY163 pKa = 10.65FDD165 pKa = 2.99QSNYY169 pKa = 10.35GGASNLFLLSPMMADD184 pKa = 3.26IAAIRR189 pKa = 11.84LLYY192 pKa = 10.85GNLATNTGDD201 pKa = 3.2TLYY204 pKa = 11.45GLGEE208 pKa = 4.3TVLGGTSDD216 pKa = 3.24MGSYY220 pKa = 10.36AGYY223 pKa = 10.83ARR225 pKa = 11.84TISDD229 pKa = 3.42SDD231 pKa = 3.67GTDD234 pKa = 2.71TMDD237 pKa = 4.05FSNVSRR243 pKa = 11.84GSVIDD248 pKa = 3.4MRR250 pKa = 11.84AGGFSSINGYY260 pKa = 9.75KK261 pKa = 10.33KK262 pKa = 10.56NFAIAQDD269 pKa = 3.76TTIEE273 pKa = 4.17NLNATLGRR281 pKa = 11.84DD282 pKa = 3.34TVTGNDD288 pKa = 4.11ADD290 pKa = 3.85NLIYY294 pKa = 10.65GLSGNDD300 pKa = 4.09KK301 pKa = 10.14ISSGAGNDD309 pKa = 3.72TLDD312 pKa = 4.03GGLGGDD318 pKa = 4.07TLTGGLGDD326 pKa = 3.86DD327 pKa = 3.7VYY329 pKa = 11.41YY330 pKa = 10.45INSSKK335 pKa = 11.13DD336 pKa = 3.59VISEE340 pKa = 4.19KK341 pKa = 10.88ADD343 pKa = 3.24GGTDD347 pKa = 3.25VVYY350 pKa = 11.02ASVSYY355 pKa = 10.59TMASYY360 pKa = 10.96LEE362 pKa = 4.21EE363 pKa = 4.1VVLQGSASTKK373 pKa = 10.62LKK375 pKa = 9.79ATGNAMANQITGDD388 pKa = 3.7DD389 pKa = 3.9GANVLNGGAGNDD401 pKa = 3.72TLAGGLGRR409 pKa = 11.84DD410 pKa = 3.7TLAGGTGNDD419 pKa = 3.83SLDD422 pKa = 3.95GGDD425 pKa = 5.51ANDD428 pKa = 4.7KK429 pKa = 10.92LDD431 pKa = 4.31GGAGDD436 pKa = 3.94DD437 pKa = 4.16TLVGGAGNDD446 pKa = 3.81LLKK449 pKa = 11.08GGAGSDD455 pKa = 2.99IFIIEE460 pKa = 5.36DD461 pKa = 3.73GTDD464 pKa = 2.94HH465 pKa = 8.02DD466 pKa = 5.18IISDD470 pKa = 3.97FAYY473 pKa = 10.5SSKK476 pKa = 9.46TSKK479 pKa = 11.08GITTIVEE486 pKa = 3.96DD487 pKa = 3.36KK488 pKa = 11.12LYY490 pKa = 9.45ITGHH494 pKa = 7.11DD495 pKa = 3.88YY496 pKa = 11.43DD497 pKa = 4.33QDD499 pKa = 3.97TLFDD503 pKa = 4.55GDD505 pKa = 4.54DD506 pKa = 3.79YY507 pKa = 11.98SVGLAADD514 pKa = 3.87DD515 pKa = 4.17SGLLFSFYY523 pKa = 10.89SGGSLLLKK531 pKa = 10.84GIGEE535 pKa = 4.35SMLDD539 pKa = 3.38KK540 pKa = 10.87FDD542 pKa = 3.53IAALHH547 pKa = 6.0TDD549 pKa = 3.76EE550 pKa = 6.1NGDD553 pKa = 3.86SYY555 pKa = 11.75FSLAA559 pKa = 4.01
Molecular weight: 58.04 kDa
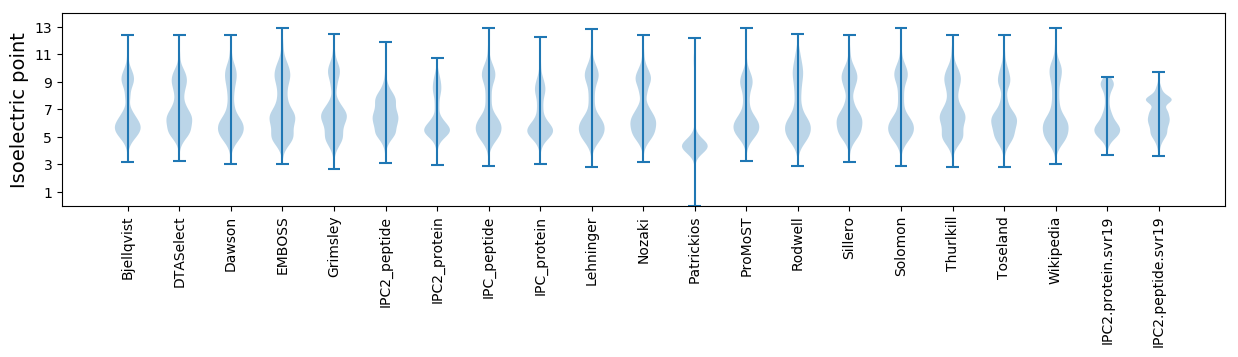
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|C6XAU7|C6XAU7_METGS Efflux transporter RND family MFP subunit OS=Methylovorus glucosetrophus (strain SIP3-4) OX=582744 GN=Msip34_2480 PE=3 SV=1
MM1 pKa = 7.21MPLHH5 pKa = 6.49RR6 pKa = 11.84PWRR9 pKa = 11.84LIRR12 pKa = 11.84GCRR15 pKa = 11.84RR16 pKa = 11.84AWLAMLGQAMLVFSMASAHH35 pKa = 6.22AAADD39 pKa = 3.46AHH41 pKa = 6.55VEE43 pKa = 3.88SHH45 pKa = 5.43YY46 pKa = 11.12RR47 pKa = 11.84EE48 pKa = 3.88AGYY51 pKa = 10.34FIGDD55 pKa = 3.82VIDD58 pKa = 3.87EE59 pKa = 4.52VVDD62 pKa = 3.51ITATPGATLVEE73 pKa = 4.38EE74 pKa = 4.22TLPRR78 pKa = 11.84SGDD81 pKa = 3.75GQWIEE86 pKa = 3.79LRR88 pKa = 11.84EE89 pKa = 4.14VNVQEE94 pKa = 4.15NAPEE98 pKa = 3.96HH99 pKa = 6.44GRR101 pKa = 11.84FKK103 pKa = 11.17LHH105 pKa = 6.99LRR107 pKa = 11.84WQLFKK112 pKa = 11.06ALRR115 pKa = 11.84EE116 pKa = 4.09TRR118 pKa = 11.84TIPLPAFTVNLRR130 pKa = 11.84HH131 pKa = 5.88GQTVLPVKK139 pKa = 9.64IHH141 pKa = 5.73EE142 pKa = 4.36RR143 pKa = 11.84QVMVSPLLPVILDD156 pKa = 3.89TSQTSLRR163 pKa = 11.84PDD165 pKa = 3.41SLPPLRR171 pKa = 11.84DD172 pKa = 3.1TLRR175 pKa = 11.84PMRR178 pKa = 11.84HH179 pKa = 4.85TVLALALTLSAALWLLWRR197 pKa = 11.84YY198 pKa = 8.71DD199 pKa = 3.55KK200 pKa = 11.4PPFRR204 pKa = 11.84WAAAGPFTAACRR216 pKa = 11.84RR217 pKa = 11.84LRR219 pKa = 11.84GRR221 pKa = 11.84QGRR224 pKa = 11.84KK225 pKa = 9.49LSLTQSFAVAHH236 pKa = 6.13HH237 pKa = 7.08AFNQAAGRR245 pKa = 11.84TVFASDD251 pKa = 3.7VPVLLKK257 pKa = 10.63LRR259 pKa = 11.84PEE261 pKa = 4.48LARR264 pKa = 11.84LRR266 pKa = 11.84PEE268 pKa = 3.54IEE270 pKa = 3.62AFYY273 pKa = 10.97ARR275 pKa = 11.84SARR278 pKa = 11.84VFFAEE283 pKa = 4.47APNAASADD291 pKa = 3.51DD292 pKa = 3.7VAAIKK297 pKa = 9.5TLLAACRR304 pKa = 11.84ALEE307 pKa = 4.13RR308 pKa = 11.84GRR310 pKa = 11.84KK311 pKa = 7.45PP312 pKa = 3.17
MM1 pKa = 7.21MPLHH5 pKa = 6.49RR6 pKa = 11.84PWRR9 pKa = 11.84LIRR12 pKa = 11.84GCRR15 pKa = 11.84RR16 pKa = 11.84AWLAMLGQAMLVFSMASAHH35 pKa = 6.22AAADD39 pKa = 3.46AHH41 pKa = 6.55VEE43 pKa = 3.88SHH45 pKa = 5.43YY46 pKa = 11.12RR47 pKa = 11.84EE48 pKa = 3.88AGYY51 pKa = 10.34FIGDD55 pKa = 3.82VIDD58 pKa = 3.87EE59 pKa = 4.52VVDD62 pKa = 3.51ITATPGATLVEE73 pKa = 4.38EE74 pKa = 4.22TLPRR78 pKa = 11.84SGDD81 pKa = 3.75GQWIEE86 pKa = 3.79LRR88 pKa = 11.84EE89 pKa = 4.14VNVQEE94 pKa = 4.15NAPEE98 pKa = 3.96HH99 pKa = 6.44GRR101 pKa = 11.84FKK103 pKa = 11.17LHH105 pKa = 6.99LRR107 pKa = 11.84WQLFKK112 pKa = 11.06ALRR115 pKa = 11.84EE116 pKa = 4.09TRR118 pKa = 11.84TIPLPAFTVNLRR130 pKa = 11.84HH131 pKa = 5.88GQTVLPVKK139 pKa = 9.64IHH141 pKa = 5.73EE142 pKa = 4.36RR143 pKa = 11.84QVMVSPLLPVILDD156 pKa = 3.89TSQTSLRR163 pKa = 11.84PDD165 pKa = 3.41SLPPLRR171 pKa = 11.84DD172 pKa = 3.1TLRR175 pKa = 11.84PMRR178 pKa = 11.84HH179 pKa = 4.85TVLALALTLSAALWLLWRR197 pKa = 11.84YY198 pKa = 8.71DD199 pKa = 3.55KK200 pKa = 11.4PPFRR204 pKa = 11.84WAAAGPFTAACRR216 pKa = 11.84RR217 pKa = 11.84LRR219 pKa = 11.84GRR221 pKa = 11.84QGRR224 pKa = 11.84KK225 pKa = 9.49LSLTQSFAVAHH236 pKa = 6.13HH237 pKa = 7.08AFNQAAGRR245 pKa = 11.84TVFASDD251 pKa = 3.7VPVLLKK257 pKa = 10.63LRR259 pKa = 11.84PEE261 pKa = 4.48LARR264 pKa = 11.84LRR266 pKa = 11.84PEE268 pKa = 3.54IEE270 pKa = 3.62AFYY273 pKa = 10.97ARR275 pKa = 11.84SARR278 pKa = 11.84VFFAEE283 pKa = 4.47APNAASADD291 pKa = 3.51DD292 pKa = 3.7VAAIKK297 pKa = 9.5TLLAACRR304 pKa = 11.84ALEE307 pKa = 4.13RR308 pKa = 11.84GRR310 pKa = 11.84KK311 pKa = 7.45PP312 pKa = 3.17
Molecular weight: 35.0 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
933751 |
23 |
2907 |
321.2 |
35.39 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
10.494 ± 0.057 | 0.822 ± 0.015 |
5.379 ± 0.036 | 5.746 ± 0.044 |
3.643 ± 0.029 | 7.297 ± 0.053 |
2.385 ± 0.029 | 5.727 ± 0.034 |
4.488 ± 0.041 | 10.682 ± 0.057 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.689 ± 0.022 | 3.623 ± 0.039 |
4.544 ± 0.033 | 4.54 ± 0.035 |
5.563 ± 0.043 | 6.004 ± 0.041 |
5.285 ± 0.042 | 6.953 ± 0.039 |
1.301 ± 0.021 | 2.837 ± 0.023 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
