
Rhodococcus sp. CUA-806
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Actinobacteria; Actinomycetia; Corynebacteriales; Nocardiaceae; Rhodococcus; unclassified Rhodococcus
Average proteome isoelectric point is 6.04
Get precalculated fractions of proteins
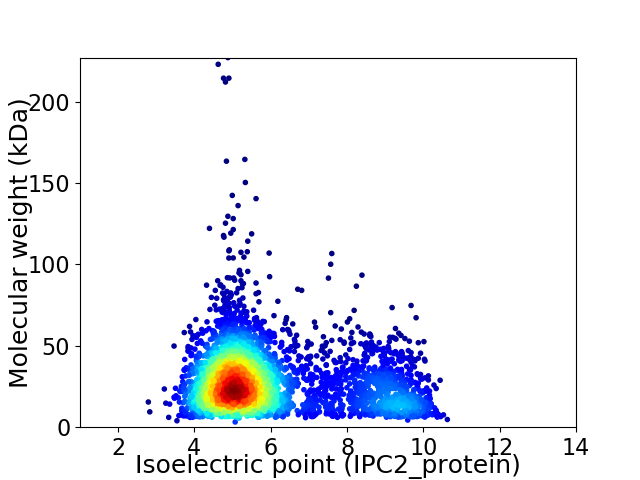
Virtual 2D-PAGE plot for 3844 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1Q9U6Q6|A0A1Q9U6Q6_9NOCA Uncharacterized protein OS=Rhodococcus sp. CUA-806 OX=1517936 GN=BJF84_22975 PE=4 SV=1
MM1 pKa = 7.44VEE3 pKa = 4.25NEE5 pKa = 4.02TPPRR9 pKa = 11.84HH10 pKa = 6.04DD11 pKa = 3.68AVMTALVIVSIVATGIIFAGLTATPEE37 pKa = 4.15EE38 pKa = 4.77PEE40 pKa = 4.39LTVQPTTTTPLPPPPQEE57 pKa = 3.74EE58 pKa = 4.86EE59 pKa = 4.56YY60 pKa = 10.82ILVTPEE66 pKa = 3.3VAYY69 pKa = 8.44PTEE72 pKa = 4.16IPGCDD77 pKa = 3.29SVEE80 pKa = 4.21EE81 pKa = 4.1PAEE84 pKa = 4.01PVYY87 pKa = 10.75SSYY90 pKa = 10.89MSDD93 pKa = 3.24GLASYY98 pKa = 10.18DD99 pKa = 3.79NPIAPWFSGPKK110 pKa = 8.74AHH112 pKa = 6.97LMSKK116 pKa = 10.62ALLASLPSDD125 pKa = 4.59LEE127 pKa = 4.3FEE129 pKa = 5.07DD130 pKa = 5.17GRR132 pKa = 11.84IPYY135 pKa = 9.41FDD137 pKa = 6.31PIPVYY142 pKa = 10.66EE143 pKa = 4.95DD144 pKa = 3.35ADD146 pKa = 3.84FTIDD150 pKa = 3.29STNAYY155 pKa = 7.65GTIGLDD161 pKa = 3.48DD162 pKa = 4.34GSGYY166 pKa = 10.7LSVGVAQFSEE176 pKa = 4.99EE177 pKa = 4.32IPPCLAGTLDD187 pKa = 3.55EE188 pKa = 5.92RR189 pKa = 11.84RR190 pKa = 11.84TQSDD194 pKa = 3.69GTVVDD199 pKa = 4.82IHH201 pKa = 5.62TTWQEE206 pKa = 3.96VNGEE210 pKa = 4.16RR211 pKa = 11.84TSTRR215 pKa = 11.84SAIAYY220 pKa = 7.56LTDD223 pKa = 4.04GSQVSAYY230 pKa = 9.12TSAIGDD236 pKa = 3.69ADD238 pKa = 4.15AVPLSVDD245 pKa = 3.21EE246 pKa = 4.39LVRR249 pKa = 11.84IVTYY253 pKa = 10.32PGLDD257 pKa = 3.29TSTAPPPGTPGNSMEE272 pKa = 4.2CTPYY276 pKa = 10.16GTDD279 pKa = 3.24TTRR282 pKa = 11.84KK283 pKa = 9.6LSPDD287 pKa = 4.23DD288 pKa = 4.18IEE290 pKa = 4.45TLNRR294 pKa = 11.84ALQQAATGGLAPSPPLGALRR314 pKa = 11.84TSSWGDD320 pKa = 3.33GLCQVVNSAAGALTVSVGGAPKK342 pKa = 9.9PDD344 pKa = 4.23APDD347 pKa = 4.15PDD349 pKa = 4.17APVPVSGPYY358 pKa = 10.45GGGTTISVPTPSGLGITVTTDD379 pKa = 4.14DD380 pKa = 3.75SWDD383 pKa = 3.52PANLEE388 pKa = 4.56RR389 pKa = 11.84IAMTPGLDD397 pKa = 3.22IPP399 pKa = 4.61
MM1 pKa = 7.44VEE3 pKa = 4.25NEE5 pKa = 4.02TPPRR9 pKa = 11.84HH10 pKa = 6.04DD11 pKa = 3.68AVMTALVIVSIVATGIIFAGLTATPEE37 pKa = 4.15EE38 pKa = 4.77PEE40 pKa = 4.39LTVQPTTTTPLPPPPQEE57 pKa = 3.74EE58 pKa = 4.86EE59 pKa = 4.56YY60 pKa = 10.82ILVTPEE66 pKa = 3.3VAYY69 pKa = 8.44PTEE72 pKa = 4.16IPGCDD77 pKa = 3.29SVEE80 pKa = 4.21EE81 pKa = 4.1PAEE84 pKa = 4.01PVYY87 pKa = 10.75SSYY90 pKa = 10.89MSDD93 pKa = 3.24GLASYY98 pKa = 10.18DD99 pKa = 3.79NPIAPWFSGPKK110 pKa = 8.74AHH112 pKa = 6.97LMSKK116 pKa = 10.62ALLASLPSDD125 pKa = 4.59LEE127 pKa = 4.3FEE129 pKa = 5.07DD130 pKa = 5.17GRR132 pKa = 11.84IPYY135 pKa = 9.41FDD137 pKa = 6.31PIPVYY142 pKa = 10.66EE143 pKa = 4.95DD144 pKa = 3.35ADD146 pKa = 3.84FTIDD150 pKa = 3.29STNAYY155 pKa = 7.65GTIGLDD161 pKa = 3.48DD162 pKa = 4.34GSGYY166 pKa = 10.7LSVGVAQFSEE176 pKa = 4.99EE177 pKa = 4.32IPPCLAGTLDD187 pKa = 3.55EE188 pKa = 5.92RR189 pKa = 11.84RR190 pKa = 11.84TQSDD194 pKa = 3.69GTVVDD199 pKa = 4.82IHH201 pKa = 5.62TTWQEE206 pKa = 3.96VNGEE210 pKa = 4.16RR211 pKa = 11.84TSTRR215 pKa = 11.84SAIAYY220 pKa = 7.56LTDD223 pKa = 4.04GSQVSAYY230 pKa = 9.12TSAIGDD236 pKa = 3.69ADD238 pKa = 4.15AVPLSVDD245 pKa = 3.21EE246 pKa = 4.39LVRR249 pKa = 11.84IVTYY253 pKa = 10.32PGLDD257 pKa = 3.29TSTAPPPGTPGNSMEE272 pKa = 4.2CTPYY276 pKa = 10.16GTDD279 pKa = 3.24TTRR282 pKa = 11.84KK283 pKa = 9.6LSPDD287 pKa = 4.23DD288 pKa = 4.18IEE290 pKa = 4.45TLNRR294 pKa = 11.84ALQQAATGGLAPSPPLGALRR314 pKa = 11.84TSSWGDD320 pKa = 3.33GLCQVVNSAAGALTVSVGGAPKK342 pKa = 9.9PDD344 pKa = 4.23APDD347 pKa = 4.15PDD349 pKa = 4.17APVPVSGPYY358 pKa = 10.45GGGTTISVPTPSGLGITVTTDD379 pKa = 4.14DD380 pKa = 3.75SWDD383 pKa = 3.52PANLEE388 pKa = 4.56RR389 pKa = 11.84IAMTPGLDD397 pKa = 3.22IPP399 pKa = 4.61
Molecular weight: 41.62 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1Q9UCV0|A0A1Q9UCV0_9NOCA 3-hydroxybutyrate dehydrogenase OS=Rhodococcus sp. CUA-806 OX=1517936 GN=BJF84_14765 PE=4 SV=1
MM1 pKa = 7.51SRR3 pKa = 11.84PLTDD7 pKa = 3.29AATSARR13 pKa = 11.84LSKK16 pKa = 10.08QRR18 pKa = 11.84RR19 pKa = 11.84RR20 pKa = 11.84DD21 pKa = 3.69TKK23 pKa = 10.6PEE25 pKa = 3.52VALRR29 pKa = 11.84RR30 pKa = 11.84EE31 pKa = 3.88LHH33 pKa = 6.02RR34 pKa = 11.84RR35 pKa = 11.84GLRR38 pKa = 11.84YY39 pKa = 9.24FVDD42 pKa = 3.95RR43 pKa = 11.84APLKK47 pKa = 10.11GMRR50 pKa = 11.84RR51 pKa = 11.84RR52 pKa = 11.84ADD54 pKa = 3.13VVFPRR59 pKa = 11.84RR60 pKa = 11.84KK61 pKa = 8.27VAVYY65 pKa = 10.57VDD67 pKa = 3.33GCFWHH72 pKa = 6.75SCPIHH77 pKa = 5.61ATQPRR82 pKa = 11.84NNAQWWADD90 pKa = 3.42KK91 pKa = 10.98LSANVARR98 pKa = 11.84DD99 pKa = 3.3RR100 pKa = 11.84NTDD103 pKa = 2.98EE104 pKa = 4.67RR105 pKa = 11.84LEE107 pKa = 4.33AEE109 pKa = 4.03GWTVVRR115 pKa = 11.84VWEE118 pKa = 4.48HH119 pKa = 5.94EE120 pKa = 4.1PAADD124 pKa = 3.53AAEE127 pKa = 4.39KK128 pKa = 10.16VAQALGG134 pKa = 3.42
MM1 pKa = 7.51SRR3 pKa = 11.84PLTDD7 pKa = 3.29AATSARR13 pKa = 11.84LSKK16 pKa = 10.08QRR18 pKa = 11.84RR19 pKa = 11.84RR20 pKa = 11.84DD21 pKa = 3.69TKK23 pKa = 10.6PEE25 pKa = 3.52VALRR29 pKa = 11.84RR30 pKa = 11.84EE31 pKa = 3.88LHH33 pKa = 6.02RR34 pKa = 11.84RR35 pKa = 11.84GLRR38 pKa = 11.84YY39 pKa = 9.24FVDD42 pKa = 3.95RR43 pKa = 11.84APLKK47 pKa = 10.11GMRR50 pKa = 11.84RR51 pKa = 11.84RR52 pKa = 11.84ADD54 pKa = 3.13VVFPRR59 pKa = 11.84RR60 pKa = 11.84KK61 pKa = 8.27VAVYY65 pKa = 10.57VDD67 pKa = 3.33GCFWHH72 pKa = 6.75SCPIHH77 pKa = 5.61ATQPRR82 pKa = 11.84NNAQWWADD90 pKa = 3.42KK91 pKa = 10.98LSANVARR98 pKa = 11.84DD99 pKa = 3.3RR100 pKa = 11.84NTDD103 pKa = 2.98EE104 pKa = 4.67RR105 pKa = 11.84LEE107 pKa = 4.33AEE109 pKa = 4.03GWTVVRR115 pKa = 11.84VWEE118 pKa = 4.48HH119 pKa = 5.94EE120 pKa = 4.1PAADD124 pKa = 3.53AAEE127 pKa = 4.39KK128 pKa = 10.16VAQALGG134 pKa = 3.42
Molecular weight: 15.5 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1045091 |
30 |
2113 |
271.9 |
29.24 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
12.321 ± 0.06 | 0.776 ± 0.011 |
6.321 ± 0.037 | 5.567 ± 0.038 |
3.101 ± 0.024 | 8.511 ± 0.036 |
2.189 ± 0.024 | 4.532 ± 0.027 |
2.214 ± 0.027 | 9.776 ± 0.041 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.056 ± 0.015 | 2.174 ± 0.02 |
5.349 ± 0.03 | 2.793 ± 0.022 |
7.155 ± 0.045 | 6.442 ± 0.03 |
6.351 ± 0.03 | 8.877 ± 0.038 |
1.452 ± 0.018 | 2.044 ± 0.017 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
