
Lachnoclostridium sp. An131
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Firmicutes; Clostridia; Eubacteriales; Lachnospiraceae; Lachnoclostridium; unclassified Lachnoclostridium
Average proteome isoelectric point is 6.02
Get precalculated fractions of proteins
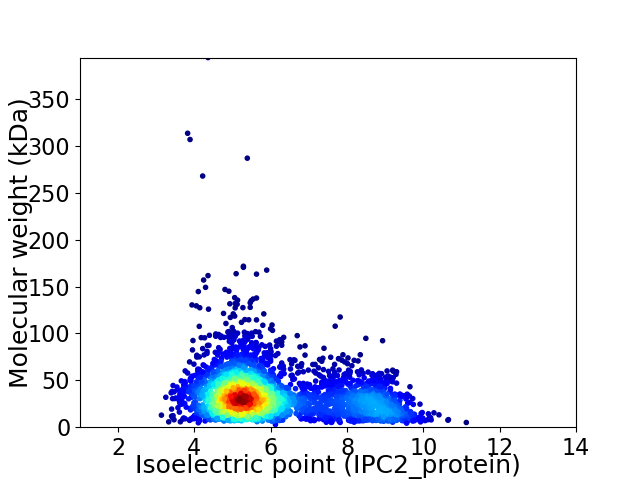
Virtual 2D-PAGE plot for 3214 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
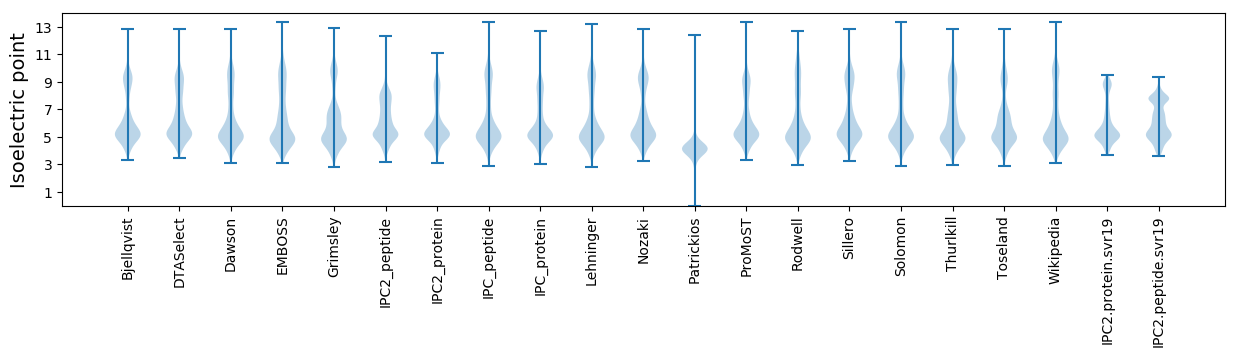
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1Y4SFH2|A0A1Y4SFH2_9FIRM Type III pantothenate kinase OS=Lachnoclostridium sp. An131 OX=1965555 GN=coaX PE=3 SV=1
MM1 pKa = 7.3KK2 pKa = 10.39RR3 pKa = 11.84KK4 pKa = 9.53LLSLTLAAVLALSLTACGGGSDD26 pKa = 3.88TADD29 pKa = 3.4TATDD33 pKa = 3.32AAEE36 pKa = 4.28TTEE39 pKa = 4.16EE40 pKa = 4.27TTEE43 pKa = 4.07EE44 pKa = 4.03TTDD47 pKa = 3.46AAEE50 pKa = 4.42STDD53 pKa = 3.5TAAGEE58 pKa = 4.16VANADD63 pKa = 3.88KK64 pKa = 10.82PLVWFNRR71 pKa = 11.84QPSNSSTGEE80 pKa = 3.83LDD82 pKa = 3.19MTALNFNEE90 pKa = 3.48NTYY93 pKa = 11.09YY94 pKa = 11.02VGFDD98 pKa = 3.41ANQGAEE104 pKa = 4.2LQGTMIRR111 pKa = 11.84DD112 pKa = 3.68YY113 pKa = 11.24IEE115 pKa = 5.38ANIDD119 pKa = 3.57TIDD122 pKa = 3.66RR123 pKa = 11.84NGDD126 pKa = 3.46GVIGYY131 pKa = 8.74VLAVGDD137 pKa = 4.11IGHH140 pKa = 6.84NDD142 pKa = 3.2SIARR146 pKa = 11.84TRR148 pKa = 11.84GVRR151 pKa = 11.84SALGTAVEE159 pKa = 4.16VDD161 pKa = 3.62GAIDD165 pKa = 3.66STPVGTNTDD174 pKa = 3.07GSSTYY179 pKa = 10.46VQDD182 pKa = 3.37GTLEE186 pKa = 4.27INGTTYY192 pKa = 9.72TVRR195 pKa = 11.84EE196 pKa = 4.12LASQEE201 pKa = 4.09MKK203 pKa = 10.84NSAGATWDD211 pKa = 3.21AATAGNAISTWSASFGDD228 pKa = 3.87QIDD231 pKa = 3.7IVASNNDD238 pKa = 3.37GMGMAMFNGWSKK250 pKa = 11.84AEE252 pKa = 3.93GVPTFGYY259 pKa = 10.15DD260 pKa = 3.27ANSDD264 pKa = 3.32AVAAIAEE271 pKa = 4.99GYY273 pKa = 10.39GGTISQHH280 pKa = 6.65ADD282 pKa = 2.86VQAYY286 pKa = 7.43LTLRR290 pKa = 11.84VLRR293 pKa = 11.84NALDD297 pKa = 3.9GVDD300 pKa = 3.46IDD302 pKa = 4.39TGIGTADD309 pKa = 3.28EE310 pKa = 4.84AGNVLTPDD318 pKa = 2.88VFTYY322 pKa = 10.9NEE324 pKa = 4.07EE325 pKa = 3.9QRR327 pKa = 11.84SYY329 pKa = 10.92YY330 pKa = 9.96ALNVAVTAEE339 pKa = 4.11NYY341 pKa = 10.16QDD343 pKa = 4.63FLDD346 pKa = 4.05STVVYY351 pKa = 10.65APVSNQLDD359 pKa = 4.15AEE361 pKa = 4.32THH363 pKa = 4.83PTKK366 pKa = 10.82NVWLNIYY373 pKa = 9.75NAADD377 pKa = 3.49NFLSATYY384 pKa = 10.31QPLLQNYY391 pKa = 9.65DD392 pKa = 3.59DD393 pKa = 4.97LLNLNVEE400 pKa = 4.63YY401 pKa = 10.44IGGDD405 pKa = 3.26GQTEE409 pKa = 4.46SNITNRR415 pKa = 11.84LGNPSQYY422 pKa = 11.16DD423 pKa = 3.19AFAINMVKK431 pKa = 9.57TDD433 pKa = 3.35NAASYY438 pKa = 10.24TSLLSQQ444 pKa = 4.08
MM1 pKa = 7.3KK2 pKa = 10.39RR3 pKa = 11.84KK4 pKa = 9.53LLSLTLAAVLALSLTACGGGSDD26 pKa = 3.88TADD29 pKa = 3.4TATDD33 pKa = 3.32AAEE36 pKa = 4.28TTEE39 pKa = 4.16EE40 pKa = 4.27TTEE43 pKa = 4.07EE44 pKa = 4.03TTDD47 pKa = 3.46AAEE50 pKa = 4.42STDD53 pKa = 3.5TAAGEE58 pKa = 4.16VANADD63 pKa = 3.88KK64 pKa = 10.82PLVWFNRR71 pKa = 11.84QPSNSSTGEE80 pKa = 3.83LDD82 pKa = 3.19MTALNFNEE90 pKa = 3.48NTYY93 pKa = 11.09YY94 pKa = 11.02VGFDD98 pKa = 3.41ANQGAEE104 pKa = 4.2LQGTMIRR111 pKa = 11.84DD112 pKa = 3.68YY113 pKa = 11.24IEE115 pKa = 5.38ANIDD119 pKa = 3.57TIDD122 pKa = 3.66RR123 pKa = 11.84NGDD126 pKa = 3.46GVIGYY131 pKa = 8.74VLAVGDD137 pKa = 4.11IGHH140 pKa = 6.84NDD142 pKa = 3.2SIARR146 pKa = 11.84TRR148 pKa = 11.84GVRR151 pKa = 11.84SALGTAVEE159 pKa = 4.16VDD161 pKa = 3.62GAIDD165 pKa = 3.66STPVGTNTDD174 pKa = 3.07GSSTYY179 pKa = 10.46VQDD182 pKa = 3.37GTLEE186 pKa = 4.27INGTTYY192 pKa = 9.72TVRR195 pKa = 11.84EE196 pKa = 4.12LASQEE201 pKa = 4.09MKK203 pKa = 10.84NSAGATWDD211 pKa = 3.21AATAGNAISTWSASFGDD228 pKa = 3.87QIDD231 pKa = 3.7IVASNNDD238 pKa = 3.37GMGMAMFNGWSKK250 pKa = 11.84AEE252 pKa = 3.93GVPTFGYY259 pKa = 10.15DD260 pKa = 3.27ANSDD264 pKa = 3.32AVAAIAEE271 pKa = 4.99GYY273 pKa = 10.39GGTISQHH280 pKa = 6.65ADD282 pKa = 2.86VQAYY286 pKa = 7.43LTLRR290 pKa = 11.84VLRR293 pKa = 11.84NALDD297 pKa = 3.9GVDD300 pKa = 3.46IDD302 pKa = 4.39TGIGTADD309 pKa = 3.28EE310 pKa = 4.84AGNVLTPDD318 pKa = 2.88VFTYY322 pKa = 10.9NEE324 pKa = 4.07EE325 pKa = 3.9QRR327 pKa = 11.84SYY329 pKa = 10.92YY330 pKa = 9.96ALNVAVTAEE339 pKa = 4.11NYY341 pKa = 10.16QDD343 pKa = 4.63FLDD346 pKa = 4.05STVVYY351 pKa = 10.65APVSNQLDD359 pKa = 4.15AEE361 pKa = 4.32THH363 pKa = 4.83PTKK366 pKa = 10.82NVWLNIYY373 pKa = 9.75NAADD377 pKa = 3.49NFLSATYY384 pKa = 10.31QPLLQNYY391 pKa = 9.65DD392 pKa = 3.59DD393 pKa = 4.97LLNLNVEE400 pKa = 4.63YY401 pKa = 10.44IGGDD405 pKa = 3.26GQTEE409 pKa = 4.46SNITNRR415 pKa = 11.84LGNPSQYY422 pKa = 11.16DD423 pKa = 3.19AFAINMVKK431 pKa = 9.57TDD433 pKa = 3.35NAASYY438 pKa = 10.24TSLLSQQ444 pKa = 4.08
Molecular weight: 47.08 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1Y4SE37|A0A1Y4SE37_9FIRM DNA topoisomerase (ATP-hydrolyzing) OS=Lachnoclostridium sp. An131 OX=1965555 GN=B5E77_08100 PE=3 SV=1
MM1 pKa = 7.67KK2 pKa = 8.72MTFQPKK8 pKa = 9.06KK9 pKa = 8.03RR10 pKa = 11.84SRR12 pKa = 11.84SKK14 pKa = 9.44VHH16 pKa = 6.17GFRR19 pKa = 11.84ARR21 pKa = 11.84MSTAGGRR28 pKa = 11.84KK29 pKa = 8.62VLAARR34 pKa = 11.84RR35 pKa = 11.84LKK37 pKa = 10.61GRR39 pKa = 11.84KK40 pKa = 8.87KK41 pKa = 10.59LSAA44 pKa = 3.95
MM1 pKa = 7.67KK2 pKa = 8.72MTFQPKK8 pKa = 9.06KK9 pKa = 8.03RR10 pKa = 11.84SRR12 pKa = 11.84SKK14 pKa = 9.44VHH16 pKa = 6.17GFRR19 pKa = 11.84ARR21 pKa = 11.84MSTAGGRR28 pKa = 11.84KK29 pKa = 8.62VLAARR34 pKa = 11.84RR35 pKa = 11.84LKK37 pKa = 10.61GRR39 pKa = 11.84KK40 pKa = 8.87KK41 pKa = 10.59LSAA44 pKa = 3.95
Molecular weight: 5.02 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1029157 |
25 |
3623 |
320.2 |
35.8 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
8.393 ± 0.053 | 1.547 ± 0.023 |
5.307 ± 0.042 | 8.327 ± 0.053 |
3.991 ± 0.031 | 7.615 ± 0.048 |
1.667 ± 0.02 | 6.4 ± 0.039 |
5.687 ± 0.042 | 9.803 ± 0.053 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.873 ± 0.023 | 3.551 ± 0.026 |
3.525 ± 0.024 | 3.35 ± 0.022 |
5.422 ± 0.04 | 5.694 ± 0.033 |
5.124 ± 0.05 | 6.747 ± 0.03 |
1.007 ± 0.014 | 3.971 ± 0.028 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
