
Xinzhou nematode virus 4
Taxonomy: Viruses; Riboviria; Orthornavirae; Negarnaviricota; Haploviricotina; Monjiviricetes; Mononegavirales; Rhabdoviridae; Alpharhabdovirinae; Alphanemrhavirus; Xinzhou alphanemrhavirus
Average proteome isoelectric point is 6.55
Get precalculated fractions of proteins
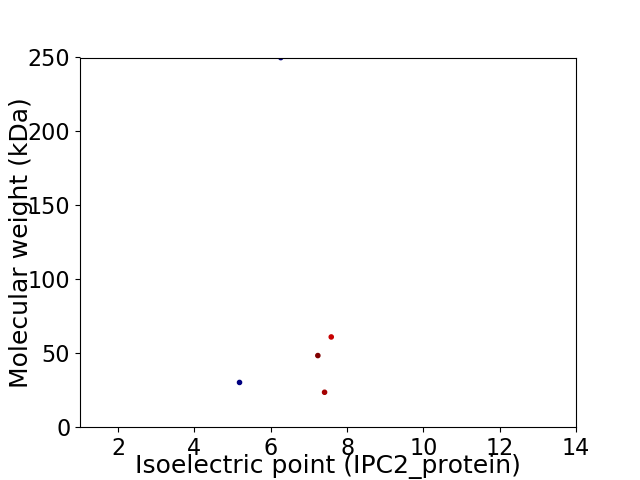
Virtual 2D-PAGE plot for 5 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
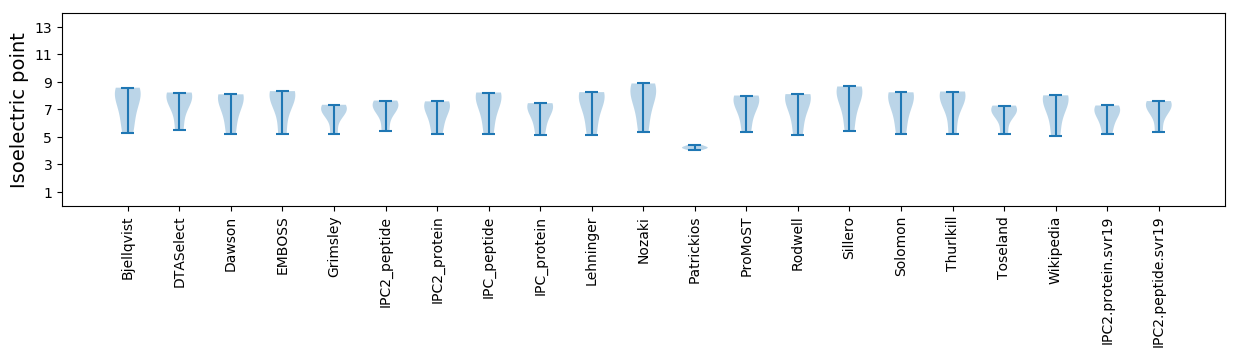
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1L3KNA4|A0A1L3KNA4_9RHAB GDP polyribonucleotidyltransferase OS=Xinzhou nematode virus 4 OX=1923772 PE=4 SV=1
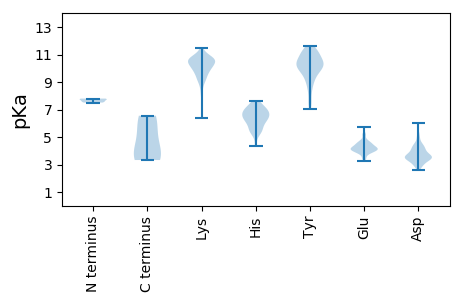
MM1 pKa = 7.69SDD3 pKa = 3.54SNEE6 pKa = 4.41SPPEE10 pKa = 3.75HH11 pKa = 6.32QFAVPAVDD19 pKa = 3.59HH20 pKa = 6.08GHH22 pKa = 7.3LARR25 pKa = 11.84LQRR28 pKa = 11.84EE29 pKa = 3.93INLRR33 pKa = 11.84ANIAQRR39 pKa = 11.84DD40 pKa = 3.87STLFRR45 pKa = 11.84HH46 pKa = 6.33VEE48 pKa = 3.98LTDD51 pKa = 4.15DD52 pKa = 3.99SPVIPTQQVFEE63 pKa = 4.68RR64 pKa = 11.84PPKK67 pKa = 10.46LSTSKK72 pKa = 8.99VTRR75 pKa = 11.84EE76 pKa = 4.2EE77 pKa = 3.87IQHH80 pKa = 6.12ARR82 pKa = 11.84TMSIISNEE90 pKa = 3.96SSKK93 pKa = 9.92TGSEE97 pKa = 4.02QQLTFPIPSTDD108 pKa = 3.88CDD110 pKa = 3.6AEE112 pKa = 4.5SYY114 pKa = 10.42IAGIHH119 pKa = 5.56AACIEE124 pKa = 4.35LTSIMKK130 pKa = 9.59FKK132 pKa = 10.41IDD134 pKa = 3.5YY135 pKa = 10.05FDD137 pKa = 5.08SNEE140 pKa = 3.71STAIVGISKK149 pKa = 8.01MHH151 pKa = 5.98SLSRR155 pKa = 11.84STSSTQMSTEE165 pKa = 4.15PLSVAPPSTQSSHH178 pKa = 5.53MDD180 pKa = 3.01VSGTEE185 pKa = 4.15EE186 pKa = 3.99PLSLNEE192 pKa = 3.45ILARR196 pKa = 11.84YY197 pKa = 8.21DD198 pKa = 3.81DD199 pKa = 4.46PVEE202 pKa = 4.09LTDD205 pKa = 4.23NLGEE209 pKa = 4.07MFQVYY214 pKa = 7.44WKK216 pKa = 11.12NEE218 pKa = 3.29ISHH221 pKa = 6.53PKK223 pKa = 9.56EE224 pKa = 3.65LLAQVIKK231 pKa = 10.86KK232 pKa = 9.64IGKK235 pKa = 9.12EE236 pKa = 3.8KK237 pKa = 10.68FLDD240 pKa = 3.77RR241 pKa = 11.84PLNQQVCLMILGDD254 pKa = 3.89PSFEE258 pKa = 4.01YY259 pKa = 10.78LRR261 pKa = 11.84GIVAPSFRR269 pKa = 11.84RR270 pKa = 3.47
MM1 pKa = 7.69SDD3 pKa = 3.54SNEE6 pKa = 4.41SPPEE10 pKa = 3.75HH11 pKa = 6.32QFAVPAVDD19 pKa = 3.59HH20 pKa = 6.08GHH22 pKa = 7.3LARR25 pKa = 11.84LQRR28 pKa = 11.84EE29 pKa = 3.93INLRR33 pKa = 11.84ANIAQRR39 pKa = 11.84DD40 pKa = 3.87STLFRR45 pKa = 11.84HH46 pKa = 6.33VEE48 pKa = 3.98LTDD51 pKa = 4.15DD52 pKa = 3.99SPVIPTQQVFEE63 pKa = 4.68RR64 pKa = 11.84PPKK67 pKa = 10.46LSTSKK72 pKa = 8.99VTRR75 pKa = 11.84EE76 pKa = 4.2EE77 pKa = 3.87IQHH80 pKa = 6.12ARR82 pKa = 11.84TMSIISNEE90 pKa = 3.96SSKK93 pKa = 9.92TGSEE97 pKa = 4.02QQLTFPIPSTDD108 pKa = 3.88CDD110 pKa = 3.6AEE112 pKa = 4.5SYY114 pKa = 10.42IAGIHH119 pKa = 5.56AACIEE124 pKa = 4.35LTSIMKK130 pKa = 9.59FKK132 pKa = 10.41IDD134 pKa = 3.5YY135 pKa = 10.05FDD137 pKa = 5.08SNEE140 pKa = 3.71STAIVGISKK149 pKa = 8.01MHH151 pKa = 5.98SLSRR155 pKa = 11.84STSSTQMSTEE165 pKa = 4.15PLSVAPPSTQSSHH178 pKa = 5.53MDD180 pKa = 3.01VSGTEE185 pKa = 4.15EE186 pKa = 3.99PLSLNEE192 pKa = 3.45ILARR196 pKa = 11.84YY197 pKa = 8.21DD198 pKa = 3.81DD199 pKa = 4.46PVEE202 pKa = 4.09LTDD205 pKa = 4.23NLGEE209 pKa = 4.07MFQVYY214 pKa = 7.44WKK216 pKa = 11.12NEE218 pKa = 3.29ISHH221 pKa = 6.53PKK223 pKa = 9.56EE224 pKa = 3.65LLAQVIKK231 pKa = 10.86KK232 pKa = 9.64IGKK235 pKa = 9.12EE236 pKa = 3.8KK237 pKa = 10.68FLDD240 pKa = 3.77RR241 pKa = 11.84PLNQQVCLMILGDD254 pKa = 3.89PSFEE258 pKa = 4.01YY259 pKa = 10.78LRR261 pKa = 11.84GIVAPSFRR269 pKa = 11.84RR270 pKa = 3.47
Molecular weight: 30.26 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1L3KNH6|A0A1L3KNH6_9RHAB Uncharacterized protein OS=Xinzhou nematode virus 4 OX=1923772 PE=4 SV=1
MM1 pKa = 7.8DD2 pKa = 3.69VSAIKK7 pKa = 10.69VLLAVLSVVHH17 pKa = 6.89PILIPDD23 pKa = 4.28CKK25 pKa = 11.02SNTVWFPATLDD36 pKa = 3.14ISMCPLPLTEE46 pKa = 4.13YY47 pKa = 9.49TSSIFKK53 pKa = 10.07EE54 pKa = 4.25GQIGLFSPEE63 pKa = 5.05DD64 pKa = 3.56MTQKK68 pKa = 10.78LVDD71 pKa = 3.63GAICYY76 pKa = 10.23IKK78 pKa = 10.49VSKK81 pKa = 10.25VKK83 pKa = 9.72CTTGFFWSQNIQKK96 pKa = 9.57SVSFRR101 pKa = 11.84RR102 pKa = 11.84PPKK105 pKa = 9.62SACRR109 pKa = 11.84DD110 pKa = 3.72HH111 pKa = 7.62IDD113 pKa = 3.36QQRR116 pKa = 11.84EE117 pKa = 4.07EE118 pKa = 4.09ISFTEE123 pKa = 4.4AEE125 pKa = 4.71FPTPVCKK132 pKa = 10.28FQLLSSTTTTTEE144 pKa = 3.77SEE146 pKa = 4.31TLIVEE151 pKa = 4.1PHH153 pKa = 6.75PIEE156 pKa = 4.11YY157 pKa = 10.16SYY159 pKa = 11.45RR160 pKa = 11.84DD161 pKa = 3.52QLLLNSVLLRR171 pKa = 11.84GKK173 pKa = 9.89CRR175 pKa = 11.84EE176 pKa = 4.36RR177 pKa = 11.84ICPTYY182 pKa = 10.47RR183 pKa = 11.84LEE185 pKa = 4.7AYY187 pKa = 8.86WIAKK191 pKa = 9.98DD192 pKa = 4.36PLPDD196 pKa = 4.17CPPMTRR202 pKa = 11.84HH203 pKa = 5.85PAKK206 pKa = 10.27IYY208 pKa = 10.57FDD210 pKa = 3.6GQKK213 pKa = 9.25PVSVISDD220 pKa = 3.46VLPPLGLKK228 pKa = 10.04GSCRR232 pKa = 11.84TVYY235 pKa = 10.26CGQPGIRR242 pKa = 11.84TVEE245 pKa = 3.97NIFINISGVDD255 pKa = 3.44ITSQIPTCTEE265 pKa = 3.49RR266 pKa = 11.84HH267 pKa = 4.44QLEE270 pKa = 4.82GFSKK274 pKa = 10.72DD275 pKa = 2.99ISSKK279 pKa = 11.3GEE281 pKa = 3.74LLTFEE286 pKa = 4.58EE287 pKa = 4.52HH288 pKa = 6.15TKK290 pKa = 9.31YY291 pKa = 9.21TLCIYY296 pKa = 10.79AVILMKK302 pKa = 10.61SLEE305 pKa = 4.3VKK307 pKa = 10.34HH308 pKa = 6.63PYY310 pKa = 9.74LLSVFQPQFAGSHH323 pKa = 5.21PCYY326 pKa = 10.21RR327 pKa = 11.84IFNRR331 pKa = 11.84TTIMKK336 pKa = 9.19AICPYY341 pKa = 10.45IHH343 pKa = 7.11MDD345 pKa = 3.46PVGVSPHH352 pKa = 5.68GVIGTDD358 pKa = 2.95KK359 pKa = 10.65HH360 pKa = 6.49SKK362 pKa = 8.29PVIWKK367 pKa = 9.36FWDD370 pKa = 3.3ALTKK374 pKa = 9.86EE375 pKa = 4.63KK376 pKa = 10.89CPSLGPNGIIRR387 pKa = 11.84DD388 pKa = 4.08EE389 pKa = 4.17QCKK392 pKa = 8.04VHH394 pKa = 6.31YY395 pKa = 8.59PHH397 pKa = 7.48EE398 pKa = 4.48YY399 pKa = 9.14VGSDD403 pKa = 2.57IMSLIGEE410 pKa = 4.56MMRR413 pKa = 11.84DD414 pKa = 3.26HH415 pKa = 7.22APIEE419 pKa = 4.0KK420 pKa = 10.54SFDD423 pKa = 2.83ILTRR427 pKa = 11.84NTQSGNVEE435 pKa = 3.55ISDD438 pKa = 3.71KK439 pKa = 10.93RR440 pKa = 11.84KK441 pKa = 9.23SQSDD445 pKa = 3.36SRR447 pKa = 11.84SFIDD451 pKa = 5.08AMSDD455 pKa = 3.75DD456 pKa = 4.18FTTLKK461 pKa = 10.77ADD463 pKa = 3.56FLIAVKK469 pKa = 10.4AFAGFCVVCVLYY481 pKa = 10.61KK482 pKa = 10.91LKK484 pKa = 10.5FFKK487 pKa = 10.95LITKK491 pKa = 7.43MLCCCISLGKK501 pKa = 10.57LSTKK505 pKa = 10.53LFVASSSTVLRR516 pKa = 11.84SSNTHH521 pKa = 5.39RR522 pKa = 11.84LRR524 pKa = 11.84RR525 pKa = 11.84RR526 pKa = 11.84QRR528 pKa = 11.84RR529 pKa = 11.84HH530 pKa = 5.35EE531 pKa = 4.56EE532 pKa = 3.5IEE534 pKa = 4.12MNPFAA539 pKa = 6.55
MM1 pKa = 7.8DD2 pKa = 3.69VSAIKK7 pKa = 10.69VLLAVLSVVHH17 pKa = 6.89PILIPDD23 pKa = 4.28CKK25 pKa = 11.02SNTVWFPATLDD36 pKa = 3.14ISMCPLPLTEE46 pKa = 4.13YY47 pKa = 9.49TSSIFKK53 pKa = 10.07EE54 pKa = 4.25GQIGLFSPEE63 pKa = 5.05DD64 pKa = 3.56MTQKK68 pKa = 10.78LVDD71 pKa = 3.63GAICYY76 pKa = 10.23IKK78 pKa = 10.49VSKK81 pKa = 10.25VKK83 pKa = 9.72CTTGFFWSQNIQKK96 pKa = 9.57SVSFRR101 pKa = 11.84RR102 pKa = 11.84PPKK105 pKa = 9.62SACRR109 pKa = 11.84DD110 pKa = 3.72HH111 pKa = 7.62IDD113 pKa = 3.36QQRR116 pKa = 11.84EE117 pKa = 4.07EE118 pKa = 4.09ISFTEE123 pKa = 4.4AEE125 pKa = 4.71FPTPVCKK132 pKa = 10.28FQLLSSTTTTTEE144 pKa = 3.77SEE146 pKa = 4.31TLIVEE151 pKa = 4.1PHH153 pKa = 6.75PIEE156 pKa = 4.11YY157 pKa = 10.16SYY159 pKa = 11.45RR160 pKa = 11.84DD161 pKa = 3.52QLLLNSVLLRR171 pKa = 11.84GKK173 pKa = 9.89CRR175 pKa = 11.84EE176 pKa = 4.36RR177 pKa = 11.84ICPTYY182 pKa = 10.47RR183 pKa = 11.84LEE185 pKa = 4.7AYY187 pKa = 8.86WIAKK191 pKa = 9.98DD192 pKa = 4.36PLPDD196 pKa = 4.17CPPMTRR202 pKa = 11.84HH203 pKa = 5.85PAKK206 pKa = 10.27IYY208 pKa = 10.57FDD210 pKa = 3.6GQKK213 pKa = 9.25PVSVISDD220 pKa = 3.46VLPPLGLKK228 pKa = 10.04GSCRR232 pKa = 11.84TVYY235 pKa = 10.26CGQPGIRR242 pKa = 11.84TVEE245 pKa = 3.97NIFINISGVDD255 pKa = 3.44ITSQIPTCTEE265 pKa = 3.49RR266 pKa = 11.84HH267 pKa = 4.44QLEE270 pKa = 4.82GFSKK274 pKa = 10.72DD275 pKa = 2.99ISSKK279 pKa = 11.3GEE281 pKa = 3.74LLTFEE286 pKa = 4.58EE287 pKa = 4.52HH288 pKa = 6.15TKK290 pKa = 9.31YY291 pKa = 9.21TLCIYY296 pKa = 10.79AVILMKK302 pKa = 10.61SLEE305 pKa = 4.3VKK307 pKa = 10.34HH308 pKa = 6.63PYY310 pKa = 9.74LLSVFQPQFAGSHH323 pKa = 5.21PCYY326 pKa = 10.21RR327 pKa = 11.84IFNRR331 pKa = 11.84TTIMKK336 pKa = 9.19AICPYY341 pKa = 10.45IHH343 pKa = 7.11MDD345 pKa = 3.46PVGVSPHH352 pKa = 5.68GVIGTDD358 pKa = 2.95KK359 pKa = 10.65HH360 pKa = 6.49SKK362 pKa = 8.29PVIWKK367 pKa = 9.36FWDD370 pKa = 3.3ALTKK374 pKa = 9.86EE375 pKa = 4.63KK376 pKa = 10.89CPSLGPNGIIRR387 pKa = 11.84DD388 pKa = 4.08EE389 pKa = 4.17QCKK392 pKa = 8.04VHH394 pKa = 6.31YY395 pKa = 8.59PHH397 pKa = 7.48EE398 pKa = 4.48YY399 pKa = 9.14VGSDD403 pKa = 2.57IMSLIGEE410 pKa = 4.56MMRR413 pKa = 11.84DD414 pKa = 3.26HH415 pKa = 7.22APIEE419 pKa = 4.0KK420 pKa = 10.54SFDD423 pKa = 2.83ILTRR427 pKa = 11.84NTQSGNVEE435 pKa = 3.55ISDD438 pKa = 3.71KK439 pKa = 10.93RR440 pKa = 11.84KK441 pKa = 9.23SQSDD445 pKa = 3.36SRR447 pKa = 11.84SFIDD451 pKa = 5.08AMSDD455 pKa = 3.75DD456 pKa = 4.18FTTLKK461 pKa = 10.77ADD463 pKa = 3.56FLIAVKK469 pKa = 10.4AFAGFCVVCVLYY481 pKa = 10.61KK482 pKa = 10.91LKK484 pKa = 10.5FFKK487 pKa = 10.95LITKK491 pKa = 7.43MLCCCISLGKK501 pKa = 10.57LSTKK505 pKa = 10.53LFVASSSTVLRR516 pKa = 11.84SSNTHH521 pKa = 5.39RR522 pKa = 11.84LRR524 pKa = 11.84RR525 pKa = 11.84RR526 pKa = 11.84QRR528 pKa = 11.84RR529 pKa = 11.84HH530 pKa = 5.35EE531 pKa = 4.56EE532 pKa = 3.5IEE534 pKa = 4.12MNPFAA539 pKa = 6.55
Molecular weight: 61.01 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
3647 |
218 |
2184 |
729.4 |
82.64 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
4.744 ± 1.103 | 2.029 ± 0.449 |
4.469 ± 0.206 | 6.115 ± 0.292 |
4.689 ± 0.37 | 5.265 ± 0.321 |
2.934 ± 0.157 | 7.211 ± 0.599 |
5.95 ± 0.348 | 11.078 ± 0.959 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.358 ± 0.105 | 4.442 ± 0.744 |
5.1 ± 0.574 | 3.427 ± 0.224 |
4.415 ± 0.276 | 9.186 ± 0.54 |
5.868 ± 0.352 | 5.703 ± 0.508 |
1.508 ± 0.359 | 3.51 ± 0.399 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
