
Saprolegnia parasitica (strain CBS 223.65)
Taxonomy: cellular organisms; Eukaryota; Sar; Stramenopiles; Oomycota; Saprolegniales; Saprolegniaceae; Saprolegnia; Saprolegnia parasitica
Average proteome isoelectric point is 6.61
Get precalculated fractions of proteins
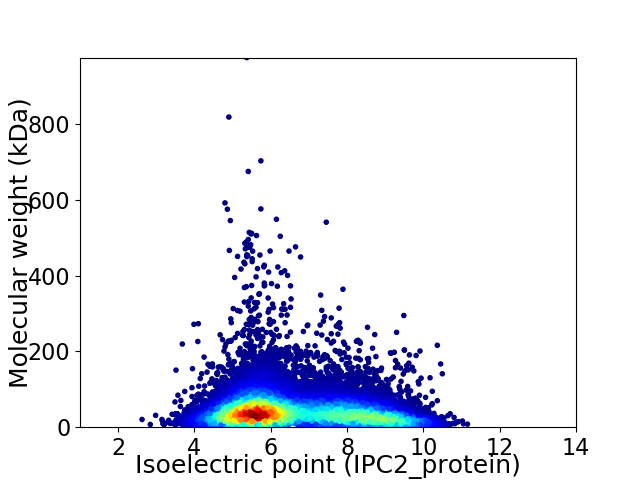
Virtual 2D-PAGE plot for 20052 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A067BWZ5|A0A067BWZ5_SAPPC Uncharacterized protein OS=Saprolegnia parasitica (strain CBS 223.65) OX=695850 GN=SPRG_12205 PE=4 SV=1
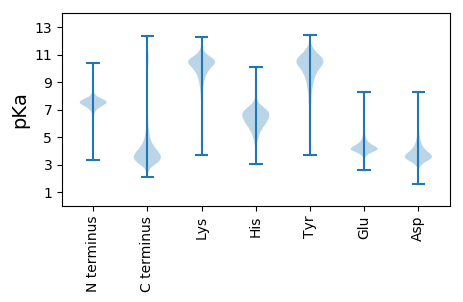
MM1 pKa = 7.09QRR3 pKa = 11.84LVCLTALLAAGAHH16 pKa = 6.53AASQPAWHH24 pKa = 6.3VCHH27 pKa = 6.44RR28 pKa = 11.84QHH30 pKa = 7.42DD31 pKa = 3.91ACATKK36 pKa = 10.65NWVCCVGPDD45 pKa = 5.08DD46 pKa = 3.83ITSGKK51 pKa = 5.0TTCRR55 pKa = 11.84PTCEE59 pKa = 4.02TTPPRR64 pKa = 11.84MKK66 pKa = 10.06LTEE69 pKa = 4.7DD70 pKa = 4.77CDD72 pKa = 5.99DD73 pKa = 6.15DD74 pKa = 7.48DD75 pKa = 7.57DD76 pKa = 7.41DD77 pKa = 6.35DD78 pKa = 6.14GNYY81 pKa = 10.7DD82 pKa = 5.56DD83 pKa = 6.7DD84 pKa = 7.41DD85 pKa = 7.12GDD87 pKa = 5.36DD88 pKa = 4.6GDD90 pKa = 5.08DD91 pKa = 3.79SGSGSVDD98 pKa = 3.3PSDD101 pKa = 5.79DD102 pKa = 3.76EE103 pKa = 5.42PSDD106 pKa = 4.2EE107 pKa = 4.96PCDD110 pKa = 4.83DD111 pKa = 4.88DD112 pKa = 6.35PSDD115 pKa = 4.08EE116 pKa = 4.83PSDD119 pKa = 3.97EE120 pKa = 4.66PSDD123 pKa = 3.99EE124 pKa = 4.66PSDD127 pKa = 3.99EE128 pKa = 4.66PSDD131 pKa = 3.99EE132 pKa = 4.66PSDD135 pKa = 3.99EE136 pKa = 4.66PSDD139 pKa = 3.99EE140 pKa = 4.66PSDD143 pKa = 3.96EE144 pKa = 4.73PSDD147 pKa = 3.99KK148 pKa = 11.03PSDD151 pKa = 3.8EE152 pKa = 4.74PSDD155 pKa = 3.8SGSGSAYY162 pKa = 8.4PTDD165 pKa = 3.9EE166 pKa = 4.92PSDD169 pKa = 4.03EE170 pKa = 4.68PSDD173 pKa = 4.12APSDD177 pKa = 4.06YY178 pKa = 10.5PSQDD182 pKa = 2.66PVYY185 pKa = 8.51PTEE188 pKa = 5.04DD189 pKa = 4.03PSPSPSPSNKK199 pKa = 7.68PTLGPRR205 pKa = 11.84PVTGDD210 pKa = 3.46LKK212 pKa = 10.19TQIIYY217 pKa = 7.06QTSVIRR223 pKa = 11.84AAHH226 pKa = 5.89GLGPVTWNDD235 pKa = 2.97EE236 pKa = 4.31LGAKK240 pKa = 7.4MQAWADD246 pKa = 3.83SNPQQNGGGHH256 pKa = 7.01GGPPGNQNLASFLVCKK272 pKa = 9.69NDD274 pKa = 3.37CMASAGPAWSWYY286 pKa = 10.12SGEE289 pKa = 4.13EE290 pKa = 4.19KK291 pKa = 10.53LWDD294 pKa = 3.55YY295 pKa = 8.86NTNKK299 pKa = 10.48SKK301 pKa = 10.99DD302 pKa = 4.11GNWMTTGHH310 pKa = 6.99FSNSMDD316 pKa = 3.67PGVNEE321 pKa = 4.05IACGYY326 pKa = 7.96STFYY330 pKa = 11.17NPTIQADD337 pKa = 4.49DD338 pKa = 3.74SLVWCNYY345 pKa = 10.3LGGNDD350 pKa = 3.68KK351 pKa = 10.69PIPRR355 pKa = 11.84PLMDD359 pKa = 4.49QEE361 pKa = 5.14ALMKK365 pKa = 10.59KK366 pKa = 10.0LVSAYY371 pKa = 10.34
MM1 pKa = 7.09QRR3 pKa = 11.84LVCLTALLAAGAHH16 pKa = 6.53AASQPAWHH24 pKa = 6.3VCHH27 pKa = 6.44RR28 pKa = 11.84QHH30 pKa = 7.42DD31 pKa = 3.91ACATKK36 pKa = 10.65NWVCCVGPDD45 pKa = 5.08DD46 pKa = 3.83ITSGKK51 pKa = 5.0TTCRR55 pKa = 11.84PTCEE59 pKa = 4.02TTPPRR64 pKa = 11.84MKK66 pKa = 10.06LTEE69 pKa = 4.7DD70 pKa = 4.77CDD72 pKa = 5.99DD73 pKa = 6.15DD74 pKa = 7.48DD75 pKa = 7.57DD76 pKa = 7.41DD77 pKa = 6.35DD78 pKa = 6.14GNYY81 pKa = 10.7DD82 pKa = 5.56DD83 pKa = 6.7DD84 pKa = 7.41DD85 pKa = 7.12GDD87 pKa = 5.36DD88 pKa = 4.6GDD90 pKa = 5.08DD91 pKa = 3.79SGSGSVDD98 pKa = 3.3PSDD101 pKa = 5.79DD102 pKa = 3.76EE103 pKa = 5.42PSDD106 pKa = 4.2EE107 pKa = 4.96PCDD110 pKa = 4.83DD111 pKa = 4.88DD112 pKa = 6.35PSDD115 pKa = 4.08EE116 pKa = 4.83PSDD119 pKa = 3.97EE120 pKa = 4.66PSDD123 pKa = 3.99EE124 pKa = 4.66PSDD127 pKa = 3.99EE128 pKa = 4.66PSDD131 pKa = 3.99EE132 pKa = 4.66PSDD135 pKa = 3.99EE136 pKa = 4.66PSDD139 pKa = 3.99EE140 pKa = 4.66PSDD143 pKa = 3.96EE144 pKa = 4.73PSDD147 pKa = 3.99KK148 pKa = 11.03PSDD151 pKa = 3.8EE152 pKa = 4.74PSDD155 pKa = 3.8SGSGSAYY162 pKa = 8.4PTDD165 pKa = 3.9EE166 pKa = 4.92PSDD169 pKa = 4.03EE170 pKa = 4.68PSDD173 pKa = 4.12APSDD177 pKa = 4.06YY178 pKa = 10.5PSQDD182 pKa = 2.66PVYY185 pKa = 8.51PTEE188 pKa = 5.04DD189 pKa = 4.03PSPSPSPSNKK199 pKa = 7.68PTLGPRR205 pKa = 11.84PVTGDD210 pKa = 3.46LKK212 pKa = 10.19TQIIYY217 pKa = 7.06QTSVIRR223 pKa = 11.84AAHH226 pKa = 5.89GLGPVTWNDD235 pKa = 2.97EE236 pKa = 4.31LGAKK240 pKa = 7.4MQAWADD246 pKa = 3.83SNPQQNGGGHH256 pKa = 7.01GGPPGNQNLASFLVCKK272 pKa = 9.69NDD274 pKa = 3.37CMASAGPAWSWYY286 pKa = 10.12SGEE289 pKa = 4.13EE290 pKa = 4.19KK291 pKa = 10.53LWDD294 pKa = 3.55YY295 pKa = 8.86NTNKK299 pKa = 10.48SKK301 pKa = 10.99DD302 pKa = 4.11GNWMTTGHH310 pKa = 6.99FSNSMDD316 pKa = 3.67PGVNEE321 pKa = 4.05IACGYY326 pKa = 7.96STFYY330 pKa = 11.17NPTIQADD337 pKa = 4.49DD338 pKa = 3.74SLVWCNYY345 pKa = 10.3LGGNDD350 pKa = 3.68KK351 pKa = 10.69PIPRR355 pKa = 11.84PLMDD359 pKa = 4.49QEE361 pKa = 5.14ALMKK365 pKa = 10.59KK366 pKa = 10.0LVSAYY371 pKa = 10.34
Molecular weight: 39.81 kDa
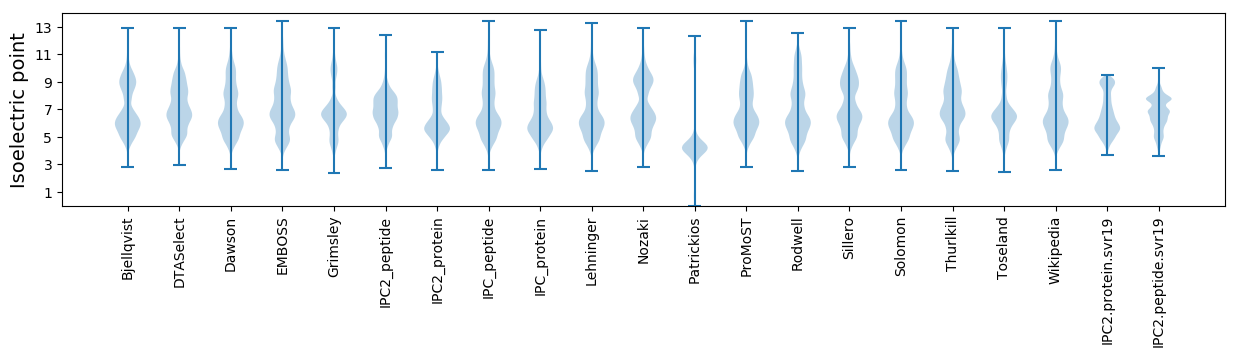
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A067BJ79|A0A067BJ79_SAPPC Uncharacterized protein OS=Saprolegnia parasitica (strain CBS 223.65) OX=695850 GN=SPRG_15383 PE=3 SV=1
MM1 pKa = 7.42APSVMSLSRR10 pKa = 11.84PTWLPAVPPRR20 pKa = 11.84AFVKK24 pKa = 10.15RR25 pKa = 11.84RR26 pKa = 11.84PRR28 pKa = 11.84PRR30 pKa = 11.84TRR32 pKa = 11.84SFAAFTPTVATTLQANTMMANGSGVTSSVGTKK64 pKa = 9.97KK65 pKa = 10.15IKK67 pKa = 10.2QPPHH71 pKa = 5.18PRR73 pKa = 11.84ARR75 pKa = 3.57
MM1 pKa = 7.42APSVMSLSRR10 pKa = 11.84PTWLPAVPPRR20 pKa = 11.84AFVKK24 pKa = 10.15RR25 pKa = 11.84RR26 pKa = 11.84PRR28 pKa = 11.84PRR30 pKa = 11.84TRR32 pKa = 11.84SFAAFTPTVATTLQANTMMANGSGVTSSVGTKK64 pKa = 9.97KK65 pKa = 10.15IKK67 pKa = 10.2QPPHH71 pKa = 5.18PRR73 pKa = 11.84ARR75 pKa = 3.57
Molecular weight: 8.16 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
8654731 |
45 |
9440 |
431.6 |
47.54 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
10.854 ± 0.02 | 1.857 ± 0.011 |
5.785 ± 0.015 | 4.959 ± 0.02 |
3.617 ± 0.012 | 5.594 ± 0.023 |
2.947 ± 0.012 | 3.961 ± 0.012 |
4.204 ± 0.021 | 10.567 ± 0.022 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.47 ± 0.008 | 2.991 ± 0.011 |
5.362 ± 0.016 | 3.684 ± 0.01 |
5.897 ± 0.02 | 7.239 ± 0.018 |
6.432 ± 0.019 | 7.468 ± 0.016 |
1.342 ± 0.006 | 2.769 ± 0.008 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
