
Taibaiella soli
Taxonomy: cellular organisms; Bacteria; FCB group; Bacteroidetes/Chlorobi group; Bacteroidetes; Chitinophagia; Chitinophagales; Chitinophagaceae; Taibaiella
Average proteome isoelectric point is 6.65
Get precalculated fractions of proteins
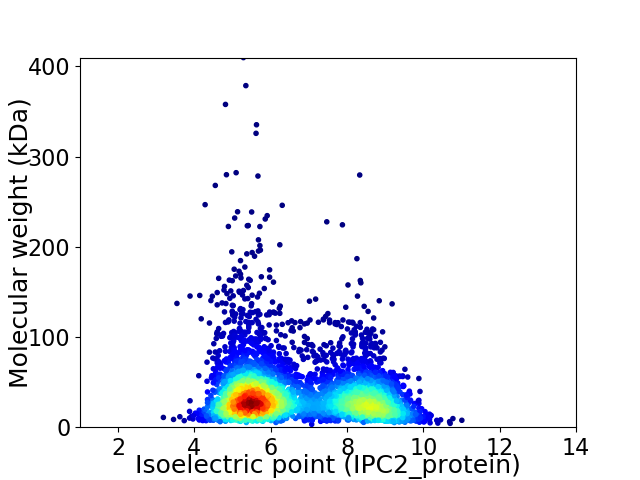
Virtual 2D-PAGE plot for 4294 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A2W2BJB6|A0A2W2BJB6_9BACT Uncharacterized protein OS=Taibaiella soli OX=1649169 GN=DN068_07365 PE=4 SV=1
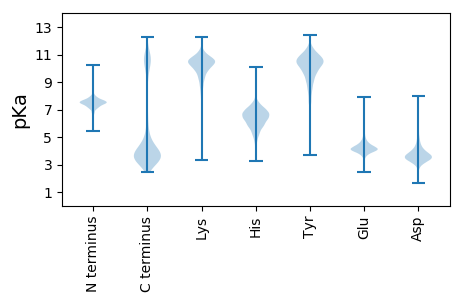
MM1 pKa = 7.27GGCVATDD8 pKa = 4.53RR9 pKa = 11.84ITVDD13 pKa = 3.56GNRR16 pKa = 11.84VGYY19 pKa = 7.82MCRR22 pKa = 11.84TEE24 pKa = 4.96PINDD28 pKa = 4.63LDD30 pKa = 4.94SGWQFLAGDD39 pKa = 4.27EE40 pKa = 4.3SQEE43 pKa = 4.18YY44 pKa = 10.93VDD46 pKa = 4.14VADD49 pKa = 3.78NSGVYY54 pKa = 9.84DD55 pKa = 3.63VNTIANYY62 pKa = 10.27DD63 pKa = 3.65PEE65 pKa = 6.15IIPFLHH71 pKa = 6.51FPIGSEE77 pKa = 4.0FEE79 pKa = 3.94RR80 pKa = 11.84DD81 pKa = 3.44EE82 pKa = 4.24VSGNLTQIRR91 pKa = 3.88
MM1 pKa = 7.27GGCVATDD8 pKa = 4.53RR9 pKa = 11.84ITVDD13 pKa = 3.56GNRR16 pKa = 11.84VGYY19 pKa = 7.82MCRR22 pKa = 11.84TEE24 pKa = 4.96PINDD28 pKa = 4.63LDD30 pKa = 4.94SGWQFLAGDD39 pKa = 4.27EE40 pKa = 4.3SQEE43 pKa = 4.18YY44 pKa = 10.93VDD46 pKa = 4.14VADD49 pKa = 3.78NSGVYY54 pKa = 9.84DD55 pKa = 3.63VNTIANYY62 pKa = 10.27DD63 pKa = 3.65PEE65 pKa = 6.15IIPFLHH71 pKa = 6.51FPIGSEE77 pKa = 4.0FEE79 pKa = 3.94RR80 pKa = 11.84DD81 pKa = 3.44EE82 pKa = 4.24VSGNLTQIRR91 pKa = 3.88
Molecular weight: 10.12 kDa
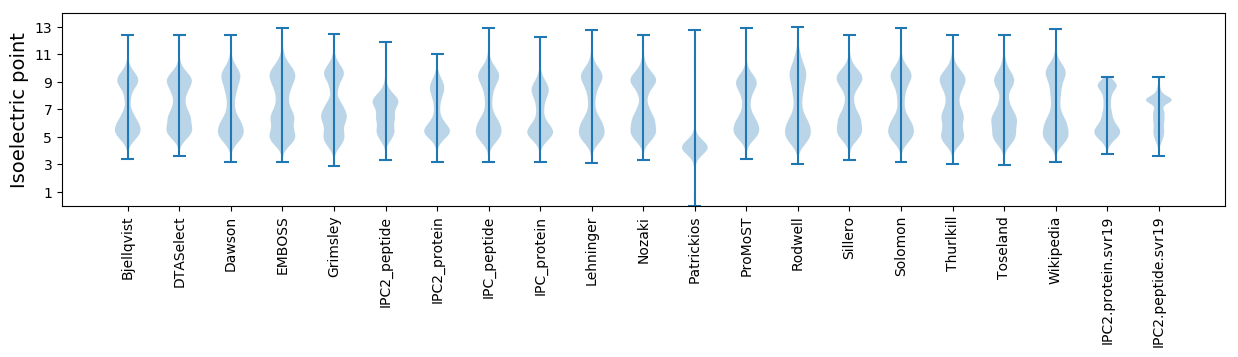
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A2W2BV27|A0A2W2BV27_9BACT Probable membrane transporter protein OS=Taibaiella soli OX=1649169 GN=DN068_16475 PE=3 SV=1
MM1 pKa = 7.64SFPSDD6 pKa = 3.47LTNSQWKK13 pKa = 9.83AISAFLPVKK22 pKa = 9.98TKK24 pKa = 10.74GIHH27 pKa = 5.38NLRR30 pKa = 11.84SIINALFYY38 pKa = 10.26IVKK41 pKa = 9.0TGCPWRR47 pKa = 11.84YY48 pKa = 9.39LPRR51 pKa = 11.84CFPLWQSVYY60 pKa = 7.84YY61 pKa = 9.94HH62 pKa = 6.23FRR64 pKa = 11.84RR65 pKa = 11.84LQSIGVLNRR74 pKa = 11.84LMSKK78 pKa = 10.11FRR80 pKa = 11.84TLLRR84 pKa = 11.84VNAGKK89 pKa = 10.35SPQPSVALIDD99 pKa = 3.79SQSIRR104 pKa = 11.84TAAGVSKK111 pKa = 10.95SKK113 pKa = 10.27GWDD116 pKa = 3.17GAKK119 pKa = 10.06RR120 pKa = 11.84LVGRR124 pKa = 11.84KK125 pKa = 7.85RR126 pKa = 11.84HH127 pKa = 5.72LVTDD131 pKa = 3.96TLGLPLALCVTSASASDD148 pKa = 3.92RR149 pKa = 11.84EE150 pKa = 4.62GMAAMTPYY158 pKa = 11.02LNRR161 pKa = 11.84IYY163 pKa = 10.82NLKK166 pKa = 8.84TLFADD171 pKa = 3.56SGYY174 pKa = 9.61MGIHH178 pKa = 5.77ITDD181 pKa = 3.34ARR183 pKa = 11.84LIITSRR189 pKa = 11.84KK190 pKa = 9.87AIADD194 pKa = 3.57KK195 pKa = 10.95CNSLSGVLTEE205 pKa = 4.67TSGTKK210 pKa = 8.76PKK212 pKa = 9.92RR213 pKa = 11.84WVVEE217 pKa = 4.61RR218 pKa = 11.84SFAWIGHH225 pKa = 4.35YY226 pKa = 9.92RR227 pKa = 11.84RR228 pKa = 11.84LAKK231 pKa = 10.54DD232 pKa = 3.76FEE234 pKa = 4.46RR235 pKa = 11.84HH236 pKa = 4.41TASSEE241 pKa = 3.76AMIKK245 pKa = 10.03IVFISIMLRR254 pKa = 11.84RR255 pKa = 11.84LNVVV259 pKa = 3.04
MM1 pKa = 7.64SFPSDD6 pKa = 3.47LTNSQWKK13 pKa = 9.83AISAFLPVKK22 pKa = 9.98TKK24 pKa = 10.74GIHH27 pKa = 5.38NLRR30 pKa = 11.84SIINALFYY38 pKa = 10.26IVKK41 pKa = 9.0TGCPWRR47 pKa = 11.84YY48 pKa = 9.39LPRR51 pKa = 11.84CFPLWQSVYY60 pKa = 7.84YY61 pKa = 9.94HH62 pKa = 6.23FRR64 pKa = 11.84RR65 pKa = 11.84LQSIGVLNRR74 pKa = 11.84LMSKK78 pKa = 10.11FRR80 pKa = 11.84TLLRR84 pKa = 11.84VNAGKK89 pKa = 10.35SPQPSVALIDD99 pKa = 3.79SQSIRR104 pKa = 11.84TAAGVSKK111 pKa = 10.95SKK113 pKa = 10.27GWDD116 pKa = 3.17GAKK119 pKa = 10.06RR120 pKa = 11.84LVGRR124 pKa = 11.84KK125 pKa = 7.85RR126 pKa = 11.84HH127 pKa = 5.72LVTDD131 pKa = 3.96TLGLPLALCVTSASASDD148 pKa = 3.92RR149 pKa = 11.84EE150 pKa = 4.62GMAAMTPYY158 pKa = 11.02LNRR161 pKa = 11.84IYY163 pKa = 10.82NLKK166 pKa = 8.84TLFADD171 pKa = 3.56SGYY174 pKa = 9.61MGIHH178 pKa = 5.77ITDD181 pKa = 3.34ARR183 pKa = 11.84LIITSRR189 pKa = 11.84KK190 pKa = 9.87AIADD194 pKa = 3.57KK195 pKa = 10.95CNSLSGVLTEE205 pKa = 4.67TSGTKK210 pKa = 8.76PKK212 pKa = 9.92RR213 pKa = 11.84WVVEE217 pKa = 4.61RR218 pKa = 11.84SFAWIGHH225 pKa = 4.35YY226 pKa = 9.92RR227 pKa = 11.84RR228 pKa = 11.84LAKK231 pKa = 10.54DD232 pKa = 3.76FEE234 pKa = 4.46RR235 pKa = 11.84HH236 pKa = 4.41TASSEE241 pKa = 3.76AMIKK245 pKa = 10.03IVFISIMLRR254 pKa = 11.84RR255 pKa = 11.84LNVVV259 pKa = 3.04
Molecular weight: 29.18 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1541204 |
26 |
3735 |
358.9 |
39.98 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.956 ± 0.042 | 1.016 ± 0.017 |
5.281 ± 0.032 | 5.055 ± 0.055 |
4.674 ± 0.028 | 7.041 ± 0.041 |
1.98 ± 0.021 | 6.768 ± 0.03 |
6.056 ± 0.051 | 8.857 ± 0.048 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.531 ± 0.02 | 5.752 ± 0.044 |
4.018 ± 0.023 | 3.953 ± 0.022 |
3.821 ± 0.034 | 6.528 ± 0.042 |
6.651 ± 0.084 | 6.576 ± 0.032 |
1.223 ± 0.013 | 4.264 ± 0.029 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
