
Halomicrobium mukohataei (strain ATCC 700874 / DSM 12286 / JCM 9738 / NCIMB 13541) (Haloarcula mukohataei)
Taxonomy: cellular organisms; Archaea; Euryarchaeota; Stenosarchaea group; Halobacteria; Halobacteriales; Haloarculaceae; Halomicrobium; Halomicrobium mukohataei
Average proteome isoelectric point is 4.89
Get precalculated fractions of proteins
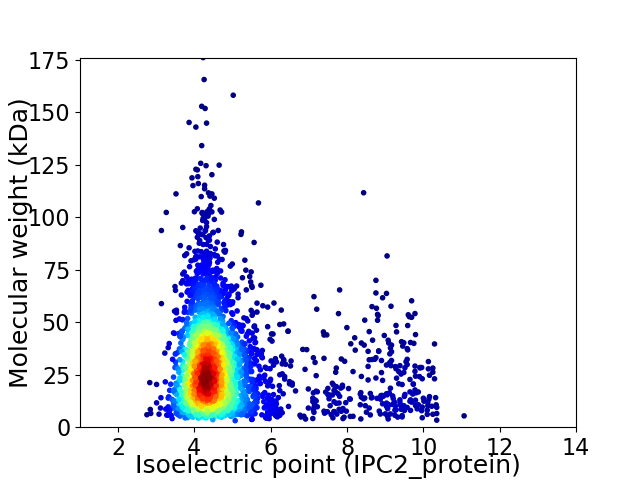
Virtual 2D-PAGE plot for 3343 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|C7P3E9|C7P3E9_HALMD Isocitrate dehydrogenase (NADP(+)) OS=Halomicrobium mukohataei (strain ATCC 700874 / DSM 12286 / JCM 9738 / NCIMB 13541) OX=485914 GN=Hmuk_1506 PE=4 SV=1
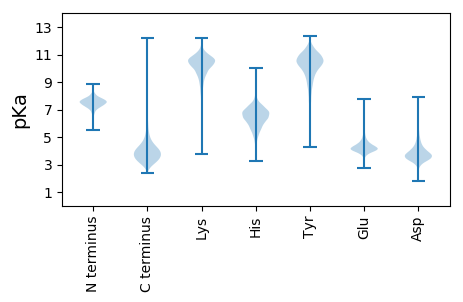
MM1 pKa = 7.98RR2 pKa = 11.84EE3 pKa = 3.53TRR5 pKa = 11.84RR6 pKa = 11.84NILRR10 pKa = 11.84KK11 pKa = 9.86ASALSALAIGVSGTASAADD30 pKa = 3.88CSDD33 pKa = 3.34VQQWQSGTAYY43 pKa = 10.5NGGDD47 pKa = 2.72RR48 pKa = 11.84VVYY51 pKa = 10.75DD52 pKa = 3.67DD53 pKa = 4.79VLWEE57 pKa = 4.22AEE59 pKa = 3.43WWTQANEE66 pKa = 4.14PEE68 pKa = 4.44EE69 pKa = 4.52SDD71 pKa = 4.81SVWTKK76 pKa = 10.72IGDD79 pKa = 3.73CSGDD83 pKa = 3.75TEE85 pKa = 4.43NEE87 pKa = 3.81APVASFTASISTPAPGEE104 pKa = 4.21SVTFDD109 pKa = 3.33ASGSSDD115 pKa = 3.34PDD117 pKa = 3.63GSVSSYY123 pKa = 11.34AWDD126 pKa = 4.12FGDD129 pKa = 4.89GDD131 pKa = 4.05TATGQTASHH140 pKa = 7.09TYY142 pKa = 10.56GSAGDD147 pKa = 3.77YY148 pKa = 9.42TVTLTVTDD156 pKa = 4.44DD157 pKa = 4.3DD158 pKa = 4.8GASGTASTTVSVSEE172 pKa = 4.78SDD174 pKa = 3.72NEE176 pKa = 3.94APNASFTVSPSSPTTGEE193 pKa = 4.16SVTVDD198 pKa = 3.12AADD201 pKa = 3.87SSDD204 pKa = 3.55ADD206 pKa = 3.92GSISSYY212 pKa = 11.3AWDD215 pKa = 4.43FGDD218 pKa = 5.12DD219 pKa = 3.6ATASGQTATHH229 pKa = 7.08TYY231 pKa = 10.24DD232 pKa = 2.94SSGEE236 pKa = 3.94YY237 pKa = 10.21TITLTVTDD245 pKa = 5.2DD246 pKa = 5.38DD247 pKa = 5.15GATDD251 pKa = 3.72TNATTVSVGGDD262 pKa = 3.06GGEE265 pKa = 4.14CSGVSEE271 pKa = 4.35WDD273 pKa = 2.99SGTTYY278 pKa = 10.47TGGDD282 pKa = 3.22QVIYY286 pKa = 10.84DD287 pKa = 3.74GTLWEE292 pKa = 4.48AKK294 pKa = 8.03WWSKK298 pKa = 10.92GDD300 pKa = 3.72EE301 pKa = 4.26PSSDD305 pKa = 3.19GGPWKK310 pKa = 10.52QITACGPPEE319 pKa = 4.42PVTKK323 pKa = 9.9TLADD327 pKa = 4.21LVPKK331 pKa = 10.43PGDD334 pKa = 3.09ITTADD339 pKa = 3.97DD340 pKa = 4.02GFEE343 pKa = 4.17ITSSTTIVAEE353 pKa = 4.42GSGTEE358 pKa = 3.93VGQYY362 pKa = 10.54LADD365 pKa = 4.01LLGPATGFDD374 pKa = 3.92LSVEE378 pKa = 4.47SGSSASADD386 pKa = 3.76SIALLLNGAPSSVGDD401 pKa = 3.63EE402 pKa = 4.45GYY404 pKa = 11.29EE405 pKa = 3.82MSVDD409 pKa = 3.45SDD411 pKa = 3.8GVTIRR416 pKa = 11.84ANEE419 pKa = 3.91AAGLFYY425 pKa = 10.75GVQSLRR431 pKa = 11.84QVLPAAVEE439 pKa = 3.94ADD441 pKa = 3.43TDD443 pKa = 3.85QSVDD447 pKa = 2.95WVVPGGSVTDD457 pKa = 3.74TPRR460 pKa = 11.84FEE462 pKa = 4.44YY463 pKa = 10.22RR464 pKa = 11.84GAMLDD469 pKa = 3.6VARR472 pKa = 11.84HH473 pKa = 5.36FFDD476 pKa = 3.39KK477 pKa = 11.08SVVKK481 pKa = 10.18EE482 pKa = 4.67FIDD485 pKa = 3.45QVAAYY490 pKa = 8.87KK491 pKa = 10.31INHH494 pKa = 6.12LHH496 pKa = 6.57LHH498 pKa = 5.7LTDD501 pKa = 3.87DD502 pKa = 3.6QGWRR506 pKa = 11.84IEE508 pKa = 3.96IDD510 pKa = 3.18DD511 pKa = 4.27WPNLTDD517 pKa = 3.55EE518 pKa = 4.88GADD521 pKa = 3.69SEE523 pKa = 4.75VDD525 pKa = 3.58GGPGGYY531 pKa = 7.59FTKK534 pKa = 10.58ADD536 pKa = 3.68YY537 pKa = 11.12QEE539 pKa = 4.75IIQYY543 pKa = 10.2AQDD546 pKa = 2.85RR547 pKa = 11.84HH548 pKa = 4.47MTVVPEE554 pKa = 3.8IDD556 pKa = 3.67MPGHH560 pKa = 6.09TGAALEE566 pKa = 4.93SYY568 pKa = 11.36AEE570 pKa = 4.37LNCDD574 pKa = 3.26DD575 pKa = 4.48TKK577 pKa = 11.11RR578 pKa = 11.84EE579 pKa = 4.0EE580 pKa = 4.05DD581 pKa = 3.1TGINVGDD588 pKa = 3.86TTLCMDD594 pKa = 5.14DD595 pKa = 3.45EE596 pKa = 4.72HH597 pKa = 8.73KK598 pKa = 9.48EE599 pKa = 4.07TSLQFAADD607 pKa = 3.93VISAVAEE614 pKa = 4.34MTDD617 pKa = 3.05GPYY620 pKa = 10.54FHH622 pKa = 7.23VGGDD626 pKa = 3.6EE627 pKa = 4.44ADD629 pKa = 3.62VLSDD633 pKa = 3.28AKK635 pKa = 11.15YY636 pKa = 10.89EE637 pKa = 4.05EE638 pKa = 5.31FIDD641 pKa = 4.24AVLPMIEE648 pKa = 4.88DD649 pKa = 3.52AGKK652 pKa = 8.33TPIGWHH658 pKa = 5.76QIASTEE664 pKa = 4.16PVTSALLHH672 pKa = 5.26YY673 pKa = 9.69WGTDD677 pKa = 3.02AQAPEE682 pKa = 3.96VAARR686 pKa = 11.84ASEE689 pKa = 4.17GNDD692 pKa = 3.18VIASPAHH699 pKa = 6.45LAYY702 pKa = 10.6LDD704 pKa = 3.38QDD706 pKa = 3.67YY707 pKa = 11.02NYY709 pKa = 10.33QDD711 pKa = 4.21GVGQDD716 pKa = 2.89WAGPVSVEE724 pKa = 5.49DD725 pKa = 4.55AYY727 pKa = 10.07TWDD730 pKa = 3.86PGSYY734 pKa = 9.22IDD736 pKa = 5.86GVDD739 pKa = 3.52EE740 pKa = 4.45SSVAGVEE747 pKa = 4.14APLWTEE753 pKa = 3.88FVEE756 pKa = 4.51TQDD759 pKa = 4.26DD760 pKa = 3.7IEE762 pKa = 5.01YY763 pKa = 9.81MVFPRR768 pKa = 11.84LAAIAEE774 pKa = 4.55LGWSSSSDD782 pKa = 2.87IGDD785 pKa = 3.66FDD787 pKa = 5.26AFSQRR792 pKa = 11.84LALQGPRR799 pKa = 11.84WAQANVNYY807 pKa = 9.74YY808 pKa = 10.54QSDD811 pKa = 3.82LVDD814 pKa = 3.41WQQ816 pKa = 3.44
MM1 pKa = 7.98RR2 pKa = 11.84EE3 pKa = 3.53TRR5 pKa = 11.84RR6 pKa = 11.84NILRR10 pKa = 11.84KK11 pKa = 9.86ASALSALAIGVSGTASAADD30 pKa = 3.88CSDD33 pKa = 3.34VQQWQSGTAYY43 pKa = 10.5NGGDD47 pKa = 2.72RR48 pKa = 11.84VVYY51 pKa = 10.75DD52 pKa = 3.67DD53 pKa = 4.79VLWEE57 pKa = 4.22AEE59 pKa = 3.43WWTQANEE66 pKa = 4.14PEE68 pKa = 4.44EE69 pKa = 4.52SDD71 pKa = 4.81SVWTKK76 pKa = 10.72IGDD79 pKa = 3.73CSGDD83 pKa = 3.75TEE85 pKa = 4.43NEE87 pKa = 3.81APVASFTASISTPAPGEE104 pKa = 4.21SVTFDD109 pKa = 3.33ASGSSDD115 pKa = 3.34PDD117 pKa = 3.63GSVSSYY123 pKa = 11.34AWDD126 pKa = 4.12FGDD129 pKa = 4.89GDD131 pKa = 4.05TATGQTASHH140 pKa = 7.09TYY142 pKa = 10.56GSAGDD147 pKa = 3.77YY148 pKa = 9.42TVTLTVTDD156 pKa = 4.44DD157 pKa = 4.3DD158 pKa = 4.8GASGTASTTVSVSEE172 pKa = 4.78SDD174 pKa = 3.72NEE176 pKa = 3.94APNASFTVSPSSPTTGEE193 pKa = 4.16SVTVDD198 pKa = 3.12AADD201 pKa = 3.87SSDD204 pKa = 3.55ADD206 pKa = 3.92GSISSYY212 pKa = 11.3AWDD215 pKa = 4.43FGDD218 pKa = 5.12DD219 pKa = 3.6ATASGQTATHH229 pKa = 7.08TYY231 pKa = 10.24DD232 pKa = 2.94SSGEE236 pKa = 3.94YY237 pKa = 10.21TITLTVTDD245 pKa = 5.2DD246 pKa = 5.38DD247 pKa = 5.15GATDD251 pKa = 3.72TNATTVSVGGDD262 pKa = 3.06GGEE265 pKa = 4.14CSGVSEE271 pKa = 4.35WDD273 pKa = 2.99SGTTYY278 pKa = 10.47TGGDD282 pKa = 3.22QVIYY286 pKa = 10.84DD287 pKa = 3.74GTLWEE292 pKa = 4.48AKK294 pKa = 8.03WWSKK298 pKa = 10.92GDD300 pKa = 3.72EE301 pKa = 4.26PSSDD305 pKa = 3.19GGPWKK310 pKa = 10.52QITACGPPEE319 pKa = 4.42PVTKK323 pKa = 9.9TLADD327 pKa = 4.21LVPKK331 pKa = 10.43PGDD334 pKa = 3.09ITTADD339 pKa = 3.97DD340 pKa = 4.02GFEE343 pKa = 4.17ITSSTTIVAEE353 pKa = 4.42GSGTEE358 pKa = 3.93VGQYY362 pKa = 10.54LADD365 pKa = 4.01LLGPATGFDD374 pKa = 3.92LSVEE378 pKa = 4.47SGSSASADD386 pKa = 3.76SIALLLNGAPSSVGDD401 pKa = 3.63EE402 pKa = 4.45GYY404 pKa = 11.29EE405 pKa = 3.82MSVDD409 pKa = 3.45SDD411 pKa = 3.8GVTIRR416 pKa = 11.84ANEE419 pKa = 3.91AAGLFYY425 pKa = 10.75GVQSLRR431 pKa = 11.84QVLPAAVEE439 pKa = 3.94ADD441 pKa = 3.43TDD443 pKa = 3.85QSVDD447 pKa = 2.95WVVPGGSVTDD457 pKa = 3.74TPRR460 pKa = 11.84FEE462 pKa = 4.44YY463 pKa = 10.22RR464 pKa = 11.84GAMLDD469 pKa = 3.6VARR472 pKa = 11.84HH473 pKa = 5.36FFDD476 pKa = 3.39KK477 pKa = 11.08SVVKK481 pKa = 10.18EE482 pKa = 4.67FIDD485 pKa = 3.45QVAAYY490 pKa = 8.87KK491 pKa = 10.31INHH494 pKa = 6.12LHH496 pKa = 6.57LHH498 pKa = 5.7LTDD501 pKa = 3.87DD502 pKa = 3.6QGWRR506 pKa = 11.84IEE508 pKa = 3.96IDD510 pKa = 3.18DD511 pKa = 4.27WPNLTDD517 pKa = 3.55EE518 pKa = 4.88GADD521 pKa = 3.69SEE523 pKa = 4.75VDD525 pKa = 3.58GGPGGYY531 pKa = 7.59FTKK534 pKa = 10.58ADD536 pKa = 3.68YY537 pKa = 11.12QEE539 pKa = 4.75IIQYY543 pKa = 10.2AQDD546 pKa = 2.85RR547 pKa = 11.84HH548 pKa = 4.47MTVVPEE554 pKa = 3.8IDD556 pKa = 3.67MPGHH560 pKa = 6.09TGAALEE566 pKa = 4.93SYY568 pKa = 11.36AEE570 pKa = 4.37LNCDD574 pKa = 3.26DD575 pKa = 4.48TKK577 pKa = 11.11RR578 pKa = 11.84EE579 pKa = 4.0EE580 pKa = 4.05DD581 pKa = 3.1TGINVGDD588 pKa = 3.86TTLCMDD594 pKa = 5.14DD595 pKa = 3.45EE596 pKa = 4.72HH597 pKa = 8.73KK598 pKa = 9.48EE599 pKa = 4.07TSLQFAADD607 pKa = 3.93VISAVAEE614 pKa = 4.34MTDD617 pKa = 3.05GPYY620 pKa = 10.54FHH622 pKa = 7.23VGGDD626 pKa = 3.6EE627 pKa = 4.44ADD629 pKa = 3.62VLSDD633 pKa = 3.28AKK635 pKa = 11.15YY636 pKa = 10.89EE637 pKa = 4.05EE638 pKa = 5.31FIDD641 pKa = 4.24AVLPMIEE648 pKa = 4.88DD649 pKa = 3.52AGKK652 pKa = 8.33TPIGWHH658 pKa = 5.76QIASTEE664 pKa = 4.16PVTSALLHH672 pKa = 5.26YY673 pKa = 9.69WGTDD677 pKa = 3.02AQAPEE682 pKa = 3.96VAARR686 pKa = 11.84ASEE689 pKa = 4.17GNDD692 pKa = 3.18VIASPAHH699 pKa = 6.45LAYY702 pKa = 10.6LDD704 pKa = 3.38QDD706 pKa = 3.67YY707 pKa = 11.02NYY709 pKa = 10.33QDD711 pKa = 4.21GVGQDD716 pKa = 2.89WAGPVSVEE724 pKa = 5.49DD725 pKa = 4.55AYY727 pKa = 10.07TWDD730 pKa = 3.86PGSYY734 pKa = 9.22IDD736 pKa = 5.86GVDD739 pKa = 3.52EE740 pKa = 4.45SSVAGVEE747 pKa = 4.14APLWTEE753 pKa = 3.88FVEE756 pKa = 4.51TQDD759 pKa = 4.26DD760 pKa = 3.7IEE762 pKa = 5.01YY763 pKa = 9.81MVFPRR768 pKa = 11.84LAAIAEE774 pKa = 4.55LGWSSSSDD782 pKa = 2.87IGDD785 pKa = 3.66FDD787 pKa = 5.26AFSQRR792 pKa = 11.84LALQGPRR799 pKa = 11.84WAQANVNYY807 pKa = 9.74YY808 pKa = 10.54QSDD811 pKa = 3.82LVDD814 pKa = 3.41WQQ816 pKa = 3.44
Molecular weight: 86.51 kDa
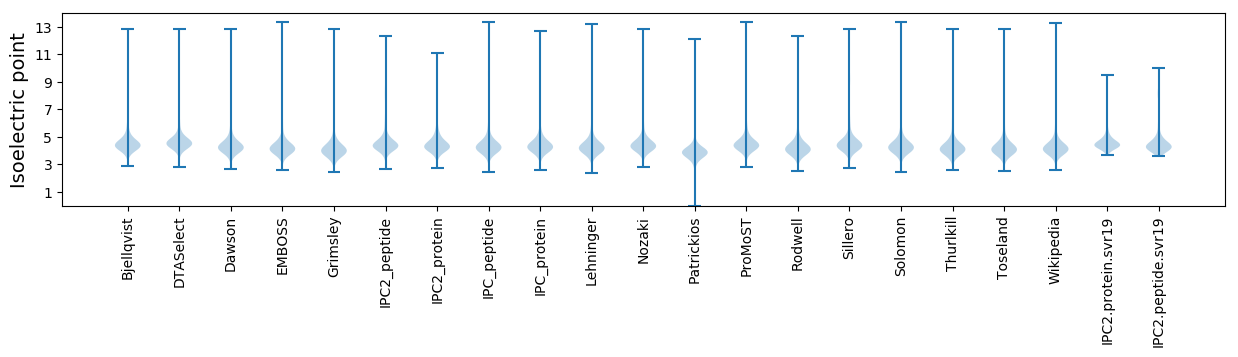
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|C7NXY6|C7NXY6_HALMD Uncharacterized protein OS=Halomicrobium mukohataei (strain ATCC 700874 / DSM 12286 / JCM 9738 / NCIMB 13541) OX=485914 GN=Hmuk_0440 PE=4 SV=1
MM1 pKa = 7.48AVLLAIATAPLTGIALVLAALIVALLARR29 pKa = 11.84AARR32 pKa = 11.84PRR34 pKa = 11.84RR35 pKa = 11.84PATSPRR41 pKa = 11.84RR42 pKa = 11.84RR43 pKa = 11.84TTVVPGTTRR52 pKa = 2.91
MM1 pKa = 7.48AVLLAIATAPLTGIALVLAALIVALLARR29 pKa = 11.84AARR32 pKa = 11.84PRR34 pKa = 11.84RR35 pKa = 11.84PATSPRR41 pKa = 11.84RR42 pKa = 11.84RR43 pKa = 11.84TTVVPGTTRR52 pKa = 2.91
Molecular weight: 5.39 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
957314 |
30 |
1589 |
286.4 |
31.07 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
11.084 ± 0.054 | 0.735 ± 0.013 |
8.656 ± 0.06 | 8.292 ± 0.053 |
3.158 ± 0.027 | 8.481 ± 0.039 |
1.972 ± 0.021 | 3.926 ± 0.032 |
1.674 ± 0.023 | 8.899 ± 0.047 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.699 ± 0.017 | 2.158 ± 0.024 |
4.616 ± 0.028 | 2.736 ± 0.03 |
6.655 ± 0.044 | 5.595 ± 0.033 |
6.677 ± 0.038 | 9.179 ± 0.045 |
1.149 ± 0.016 | 2.66 ± 0.024 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
