
Youngiibacter fragilis 232.1
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Firmicutes; Clostridia; Eubacteriales; Clostridiaceae; Youngiibacter; Youngiibacter fragilis
Average proteome isoelectric point is 6.2
Get precalculated fractions of proteins
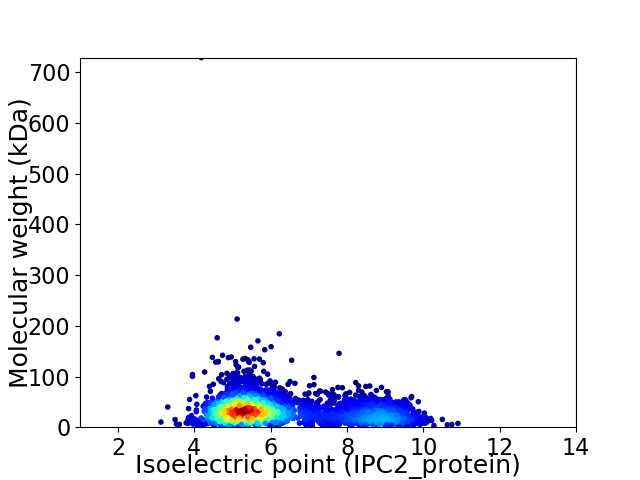
Virtual 2D-PAGE plot for 3729 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|V7I4L7|V7I4L7_9CLOT 50S ribosomal protein L35 OS=Youngiibacter fragilis 232.1 OX=994573 GN=rpmI PE=3 SV=1
MM1 pKa = 7.51IVTGSSIIQPTSLTLLPSAITEE23 pKa = 3.67IHH25 pKa = 6.34PTDD28 pKa = 4.77LYY30 pKa = 10.07PTVLTIDD37 pKa = 3.54SVTPADD43 pKa = 4.63ADD45 pKa = 3.6LTDD48 pKa = 4.44IYY50 pKa = 10.29WISTDD55 pKa = 3.43PSVATVVKK63 pKa = 9.76TGLLTADD70 pKa = 3.58VAAKK74 pKa = 10.47SNGYY78 pKa = 9.68AAIQVFRR85 pKa = 11.84NDD87 pKa = 3.14GLFLGSCPVTVTTDD101 pKa = 3.12EE102 pKa = 4.35TLTDD106 pKa = 3.7PVYY109 pKa = 10.74IQATDD114 pKa = 3.77FNGVPLDD121 pKa = 3.66QFPDD125 pKa = 3.39KK126 pKa = 11.08DD127 pKa = 4.03SIYY130 pKa = 8.8ITCYY134 pKa = 10.89NLDD137 pKa = 3.83THH139 pKa = 7.14PGPYY143 pKa = 9.98AIRR146 pKa = 11.84VSEE149 pKa = 4.22KK150 pKa = 10.71SSDD153 pKa = 4.02PILGMNLIDD162 pKa = 3.97GVKK165 pKa = 10.44DD166 pKa = 3.35LVISEE171 pKa = 4.3GQSTYY176 pKa = 8.01MFRR179 pKa = 11.84LIDD182 pKa = 3.78YY183 pKa = 10.89VDD185 pKa = 5.24FEE187 pKa = 4.58PTTNFSKK194 pKa = 11.31SNFVSMSLSDD204 pKa = 3.87NFPTGDD210 pKa = 4.04YY211 pKa = 11.09EE212 pKa = 6.03DD213 pKa = 4.47GTAQTLTDD221 pKa = 3.75NFKK224 pKa = 9.48VTSPVPTGDD233 pKa = 2.79IDD235 pKa = 3.86VDD237 pKa = 3.54IRR239 pKa = 11.84QLDD242 pKa = 3.76QFDD245 pKa = 3.71KK246 pKa = 11.18ASAYY250 pKa = 9.03LHH252 pKa = 6.28PSIIGMNVILGRR264 pKa = 11.84EE265 pKa = 4.17IKK267 pKa = 10.45DD268 pKa = 3.26QTVLEE273 pKa = 4.46TTYY276 pKa = 11.51EE277 pKa = 4.25DD278 pKa = 4.18YY279 pKa = 11.46LNPAWYY285 pKa = 9.15EE286 pKa = 4.1GAPSTIPKK294 pKa = 8.54YY295 pKa = 9.14TDD297 pKa = 3.0EE298 pKa = 4.94AKK300 pKa = 10.93LIGQVADD307 pKa = 3.21VDD309 pKa = 4.37LDD311 pKa = 3.48GDD313 pKa = 4.8YY314 pKa = 10.7EE315 pKa = 4.56VHH317 pKa = 6.47WNPPKK322 pKa = 10.66EE323 pKa = 3.86PLNIGGYY330 pKa = 9.69VLLII334 pKa = 4.1
MM1 pKa = 7.51IVTGSSIIQPTSLTLLPSAITEE23 pKa = 3.67IHH25 pKa = 6.34PTDD28 pKa = 4.77LYY30 pKa = 10.07PTVLTIDD37 pKa = 3.54SVTPADD43 pKa = 4.63ADD45 pKa = 3.6LTDD48 pKa = 4.44IYY50 pKa = 10.29WISTDD55 pKa = 3.43PSVATVVKK63 pKa = 9.76TGLLTADD70 pKa = 3.58VAAKK74 pKa = 10.47SNGYY78 pKa = 9.68AAIQVFRR85 pKa = 11.84NDD87 pKa = 3.14GLFLGSCPVTVTTDD101 pKa = 3.12EE102 pKa = 4.35TLTDD106 pKa = 3.7PVYY109 pKa = 10.74IQATDD114 pKa = 3.77FNGVPLDD121 pKa = 3.66QFPDD125 pKa = 3.39KK126 pKa = 11.08DD127 pKa = 4.03SIYY130 pKa = 8.8ITCYY134 pKa = 10.89NLDD137 pKa = 3.83THH139 pKa = 7.14PGPYY143 pKa = 9.98AIRR146 pKa = 11.84VSEE149 pKa = 4.22KK150 pKa = 10.71SSDD153 pKa = 4.02PILGMNLIDD162 pKa = 3.97GVKK165 pKa = 10.44DD166 pKa = 3.35LVISEE171 pKa = 4.3GQSTYY176 pKa = 8.01MFRR179 pKa = 11.84LIDD182 pKa = 3.78YY183 pKa = 10.89VDD185 pKa = 5.24FEE187 pKa = 4.58PTTNFSKK194 pKa = 11.31SNFVSMSLSDD204 pKa = 3.87NFPTGDD210 pKa = 4.04YY211 pKa = 11.09EE212 pKa = 6.03DD213 pKa = 4.47GTAQTLTDD221 pKa = 3.75NFKK224 pKa = 9.48VTSPVPTGDD233 pKa = 2.79IDD235 pKa = 3.86VDD237 pKa = 3.54IRR239 pKa = 11.84QLDD242 pKa = 3.76QFDD245 pKa = 3.71KK246 pKa = 11.18ASAYY250 pKa = 9.03LHH252 pKa = 6.28PSIIGMNVILGRR264 pKa = 11.84EE265 pKa = 4.17IKK267 pKa = 10.45DD268 pKa = 3.26QTVLEE273 pKa = 4.46TTYY276 pKa = 11.51EE277 pKa = 4.25DD278 pKa = 4.18YY279 pKa = 11.46LNPAWYY285 pKa = 9.15EE286 pKa = 4.1GAPSTIPKK294 pKa = 8.54YY295 pKa = 9.14TDD297 pKa = 3.0EE298 pKa = 4.94AKK300 pKa = 10.93LIGQVADD307 pKa = 3.21VDD309 pKa = 4.37LDD311 pKa = 3.48GDD313 pKa = 4.8YY314 pKa = 10.7EE315 pKa = 4.56VHH317 pKa = 6.47WNPPKK322 pKa = 10.66EE323 pKa = 3.86PLNIGGYY330 pKa = 9.69VLLII334 pKa = 4.1
Molecular weight: 36.58 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|V7HZJ8|V7HZJ8_9CLOT Mannose-6-phosphate isomerase OS=Youngiibacter fragilis 232.1 OX=994573 GN=T472_0218865 PE=4 SV=1
MM1 pKa = 7.81KK2 pKa = 10.19EE3 pKa = 3.82RR4 pKa = 11.84LRR6 pKa = 11.84KK7 pKa = 9.83LMQGRR12 pKa = 11.84YY13 pKa = 9.55GVDD16 pKa = 2.83EE17 pKa = 4.8LSRR20 pKa = 11.84LMVYY24 pKa = 10.41VSFVVMLIGSFAKK37 pKa = 10.26NPYY40 pKa = 9.11INLVGFAMTIYY51 pKa = 9.83AYY53 pKa = 10.93SRR55 pKa = 11.84IFSKK59 pKa = 9.19NHH61 pKa = 6.19RR62 pKa = 11.84LCSSQNLKK70 pKa = 10.21YY71 pKa = 10.65VQVRR75 pKa = 11.84DD76 pKa = 3.22RR77 pKa = 11.84FLRR80 pKa = 11.84KK81 pKa = 9.02IANHH85 pKa = 4.73VQIMKK90 pKa = 10.56LSRR93 pKa = 11.84TYY95 pKa = 10.29RR96 pKa = 11.84VYY98 pKa = 10.78SCPGCRR104 pKa = 11.84QMVRR108 pKa = 11.84IPKK111 pKa = 10.06GKK113 pKa = 10.08GKK115 pKa = 10.75VEE117 pKa = 4.22VKK119 pKa = 10.4CPKK122 pKa = 10.02CGTRR126 pKa = 11.84FSRR129 pKa = 11.84KK130 pKa = 7.0SS131 pKa = 3.12
MM1 pKa = 7.81KK2 pKa = 10.19EE3 pKa = 3.82RR4 pKa = 11.84LRR6 pKa = 11.84KK7 pKa = 9.83LMQGRR12 pKa = 11.84YY13 pKa = 9.55GVDD16 pKa = 2.83EE17 pKa = 4.8LSRR20 pKa = 11.84LMVYY24 pKa = 10.41VSFVVMLIGSFAKK37 pKa = 10.26NPYY40 pKa = 9.11INLVGFAMTIYY51 pKa = 9.83AYY53 pKa = 10.93SRR55 pKa = 11.84IFSKK59 pKa = 9.19NHH61 pKa = 6.19RR62 pKa = 11.84LCSSQNLKK70 pKa = 10.21YY71 pKa = 10.65VQVRR75 pKa = 11.84DD76 pKa = 3.22RR77 pKa = 11.84FLRR80 pKa = 11.84KK81 pKa = 9.02IANHH85 pKa = 4.73VQIMKK90 pKa = 10.56LSRR93 pKa = 11.84TYY95 pKa = 10.29RR96 pKa = 11.84VYY98 pKa = 10.78SCPGCRR104 pKa = 11.84QMVRR108 pKa = 11.84IPKK111 pKa = 10.06GKK113 pKa = 10.08GKK115 pKa = 10.75VEE117 pKa = 4.22VKK119 pKa = 10.4CPKK122 pKa = 10.02CGTRR126 pKa = 11.84FSRR129 pKa = 11.84KK130 pKa = 7.0SS131 pKa = 3.12
Molecular weight: 15.36 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1132455 |
29 |
7056 |
303.7 |
33.78 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.561 ± 0.049 | 0.995 ± 0.016 |
5.784 ± 0.034 | 7.315 ± 0.047 |
4.161 ± 0.031 | 7.818 ± 0.044 |
1.626 ± 0.019 | 7.667 ± 0.037 |
6.574 ± 0.038 | 9.296 ± 0.041 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
3.214 ± 0.023 | 3.993 ± 0.022 |
3.443 ± 0.022 | 2.242 ± 0.019 |
5.023 ± 0.044 | 6.532 ± 0.034 |
5.258 ± 0.033 | 7.112 ± 0.035 |
0.779 ± 0.011 | 3.608 ± 0.024 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
