
Eimeria tenella (Coccidian parasite)
Taxonomy: cellular organisms; Eukaryota; Sar; Alveolata; Apicomplexa; Conoidasida; Coccidia; Eucoccidiorida; Eimeriorina; Eimeriidae; Eimeria
Average proteome isoelectric point is 6.95
Get precalculated fractions of proteins
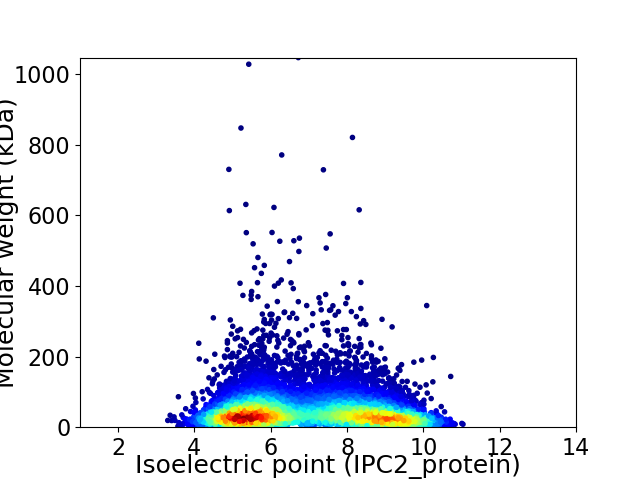
Virtual 2D-PAGE plot for 8595 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|U6KV80|U6KV80_EIMTE Uncharacterized protein OS=Eimeria tenella OX=5802 GN=ETH_00020980 PE=4 SV=1
MM1 pKa = 7.08EE2 pKa = 5.34AAAATDD8 pKa = 3.57TAAAAAATGVAAATAAEE25 pKa = 4.49VTTEE29 pKa = 3.73AAAEE33 pKa = 4.25TSCAAVQAAQATFAVSGAAASAADD57 pKa = 3.53EE58 pKa = 4.31AAANRR63 pKa = 11.84IDD65 pKa = 3.85FVRR68 pKa = 11.84AADD71 pKa = 3.71TAAAAAAADD80 pKa = 4.08ADD82 pKa = 3.92AQKK85 pKa = 10.96PIMKK89 pKa = 9.66EE90 pKa = 4.17LYY92 pKa = 10.33ASAAAAADD100 pKa = 3.53AATAGIAADD109 pKa = 3.94AAGEE113 pKa = 4.06AADD116 pKa = 4.21AAAGAADD123 pKa = 3.99AAAAAADD130 pKa = 3.84AAADD134 pKa = 3.48AAAAADD140 pKa = 3.66ASTEE144 pKa = 3.97TSAAATDD151 pKa = 3.98TSSLGAAAADD161 pKa = 3.68VAAADD166 pKa = 3.89LAAAAEE172 pKa = 4.32VATAEE177 pKa = 4.25LAEE180 pKa = 4.37AAGDD184 pKa = 3.97PADD187 pKa = 4.1AAADD191 pKa = 3.59AAAAAAEE198 pKa = 4.32AACDD202 pKa = 3.59ACNFCADD209 pKa = 3.84FCSNSWCLRR218 pKa = 11.84CLPLRR223 pKa = 11.84QQTRR227 pKa = 11.84EE228 pKa = 4.11RR229 pKa = 11.84LWGPCCCSCCSCCFCNTAAAAEE251 pKa = 4.0AAAAAEE257 pKa = 4.38AGFAAEE263 pKa = 4.89PNSSSIDD270 pKa = 3.57SYY272 pKa = 11.25NAAVAAAAALAADD285 pKa = 4.18RR286 pKa = 11.84RR287 pKa = 11.84RR288 pKa = 11.84RR289 pKa = 11.84GG290 pKa = 3.1
MM1 pKa = 7.08EE2 pKa = 5.34AAAATDD8 pKa = 3.57TAAAAAATGVAAATAAEE25 pKa = 4.49VTTEE29 pKa = 3.73AAAEE33 pKa = 4.25TSCAAVQAAQATFAVSGAAASAADD57 pKa = 3.53EE58 pKa = 4.31AAANRR63 pKa = 11.84IDD65 pKa = 3.85FVRR68 pKa = 11.84AADD71 pKa = 3.71TAAAAAAADD80 pKa = 4.08ADD82 pKa = 3.92AQKK85 pKa = 10.96PIMKK89 pKa = 9.66EE90 pKa = 4.17LYY92 pKa = 10.33ASAAAAADD100 pKa = 3.53AATAGIAADD109 pKa = 3.94AAGEE113 pKa = 4.06AADD116 pKa = 4.21AAAGAADD123 pKa = 3.99AAAAAADD130 pKa = 3.84AAADD134 pKa = 3.48AAAAADD140 pKa = 3.66ASTEE144 pKa = 3.97TSAAATDD151 pKa = 3.98TSSLGAAAADD161 pKa = 3.68VAAADD166 pKa = 3.89LAAAAEE172 pKa = 4.32VATAEE177 pKa = 4.25LAEE180 pKa = 4.37AAGDD184 pKa = 3.97PADD187 pKa = 4.1AAADD191 pKa = 3.59AAAAAAEE198 pKa = 4.32AACDD202 pKa = 3.59ACNFCADD209 pKa = 3.84FCSNSWCLRR218 pKa = 11.84CLPLRR223 pKa = 11.84QQTRR227 pKa = 11.84EE228 pKa = 4.11RR229 pKa = 11.84LWGPCCCSCCSCCFCNTAAAAEE251 pKa = 4.0AAAAAEE257 pKa = 4.38AGFAAEE263 pKa = 4.89PNSSSIDD270 pKa = 3.57SYY272 pKa = 11.25NAAVAAAAALAADD285 pKa = 4.18RR286 pKa = 11.84RR287 pKa = 11.84RR288 pKa = 11.84RR289 pKa = 11.84GG290 pKa = 3.1
Molecular weight: 27.08 kDa
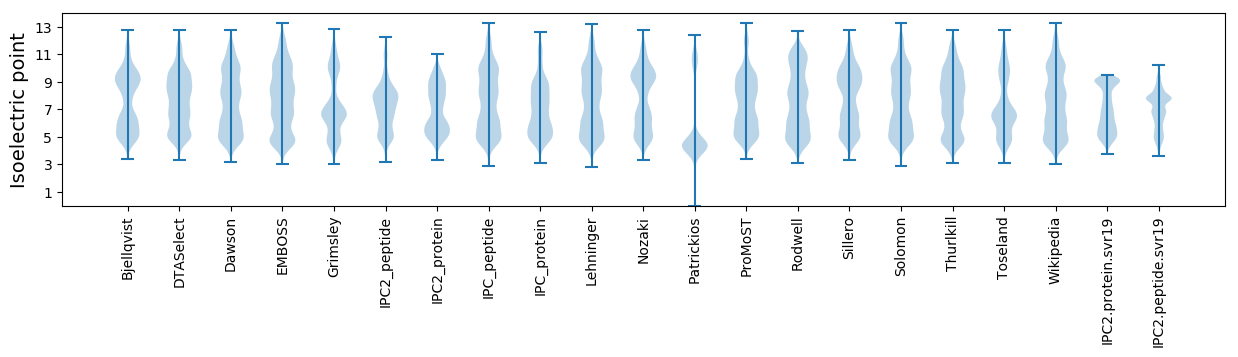
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|U6KV52|U6KV52_EIMTE Rna polymerase ii b3 subunit OS=Eimeria tenella OX=5802 GN=ETH_00035430 PE=4 SV=1
RR1 pKa = 6.82PTALVLPFGTHH12 pKa = 5.75SSSSSSNSSSSSSSKK27 pKa = 10.39NCSSTSSRR35 pKa = 11.84RR36 pKa = 11.84VKK38 pKa = 10.26RR39 pKa = 11.84MRR41 pKa = 11.84MKK43 pKa = 10.53RR44 pKa = 11.84RR45 pKa = 11.84RR46 pKa = 11.84SGKK49 pKa = 9.51NSSKK53 pKa = 9.69TSSSSSSKK61 pKa = 9.47SSRR64 pKa = 11.84RR65 pKa = 11.84SRR67 pKa = 11.84SKK69 pKa = 9.87RR70 pKa = 11.84SSSRR74 pKa = 11.84RR75 pKa = 11.84SSSS78 pKa = 2.78
RR1 pKa = 6.82PTALVLPFGTHH12 pKa = 5.75SSSSSSNSSSSSSSKK27 pKa = 10.39NCSSTSSRR35 pKa = 11.84RR36 pKa = 11.84VKK38 pKa = 10.26RR39 pKa = 11.84MRR41 pKa = 11.84MKK43 pKa = 10.53RR44 pKa = 11.84RR45 pKa = 11.84RR46 pKa = 11.84SGKK49 pKa = 9.51NSSKK53 pKa = 9.69TSSSSSSKK61 pKa = 9.47SSRR64 pKa = 11.84RR65 pKa = 11.84SRR67 pKa = 11.84SKK69 pKa = 9.87RR70 pKa = 11.84SSSRR74 pKa = 11.84RR75 pKa = 11.84SSSS78 pKa = 2.78
Molecular weight: 8.44 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
4341326 |
10 |
9860 |
505.1 |
54.22 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
14.753 ± 0.089 | 2.015 ± 0.017 |
3.612 ± 0.021 | 6.173 ± 0.03 |
2.743 ± 0.018 | 6.457 ± 0.031 |
2.07 ± 0.011 | 2.431 ± 0.021 |
3.794 ± 0.029 | 10.069 ± 0.043 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.387 ± 0.011 | 2.229 ± 0.013 |
6.093 ± 0.032 | 9.034 ± 0.079 |
6.151 ± 0.025 | 9.219 ± 0.045 |
4.341 ± 0.022 | 4.956 ± 0.025 |
1.0 ± 0.007 | 1.472 ± 0.012 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
