
Gammapapillomavirus 11
Taxonomy: Viruses; Monodnaviria; Shotokuvirae; Cossaviricota; Papovaviricetes; Zurhausenvirales; Papillomaviridae; Firstpapillomavirinae; Gammapapillomavirus
Average proteome isoelectric point is 6.48
Get precalculated fractions of proteins
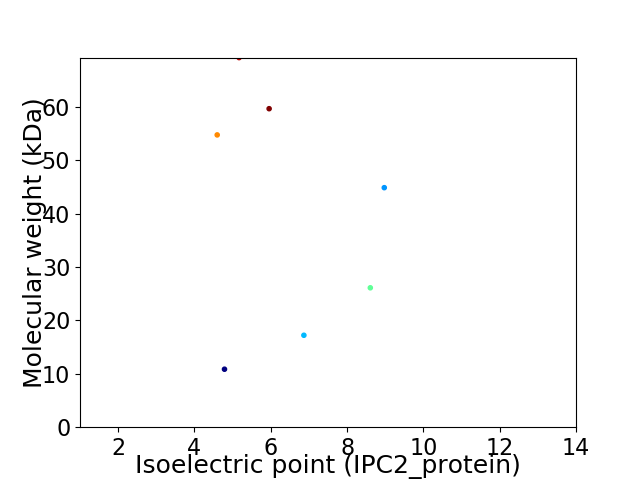
Virtual 2D-PAGE plot for 7 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A2D2ALL3|A0A2D2ALL3_9PAPI Regulatory protein E2 OS=Gammapapillomavirus 11 OX=1513256 GN=E2 PE=3 SV=1
MM1 pKa = 7.11MGEE4 pKa = 4.45KK5 pKa = 8.75PTLKK9 pKa = 10.58DD10 pKa = 3.0IVLEE14 pKa = 4.05EE15 pKa = 4.0QLADD19 pKa = 3.3MVMPANLLCEE29 pKa = 4.45EE30 pKa = 4.69SLSPDD35 pKa = 3.34DD36 pKa = 4.0TPEE39 pKa = 4.08EE40 pKa = 4.34EE41 pKa = 4.64SLSPYY46 pKa = 10.36QVDD49 pKa = 4.36SLCKK53 pKa = 9.33ACNKK57 pKa = 9.64RR58 pKa = 11.84VRR60 pKa = 11.84LFVVATAGAIFVLEE74 pKa = 3.74QLLFQNLSIVCPVCSRR90 pKa = 11.84SNLHH94 pKa = 6.74HH95 pKa = 6.39GRR97 pKa = 11.84SS98 pKa = 3.64
MM1 pKa = 7.11MGEE4 pKa = 4.45KK5 pKa = 8.75PTLKK9 pKa = 10.58DD10 pKa = 3.0IVLEE14 pKa = 4.05EE15 pKa = 4.0QLADD19 pKa = 3.3MVMPANLLCEE29 pKa = 4.45EE30 pKa = 4.69SLSPDD35 pKa = 3.34DD36 pKa = 4.0TPEE39 pKa = 4.08EE40 pKa = 4.34EE41 pKa = 4.64SLSPYY46 pKa = 10.36QVDD49 pKa = 4.36SLCKK53 pKa = 9.33ACNKK57 pKa = 9.64RR58 pKa = 11.84VRR60 pKa = 11.84LFVVATAGAIFVLEE74 pKa = 3.74QLLFQNLSIVCPVCSRR90 pKa = 11.84SNLHH94 pKa = 6.74HH95 pKa = 6.39GRR97 pKa = 11.84SS98 pKa = 3.64
Molecular weight: 10.86 kDa
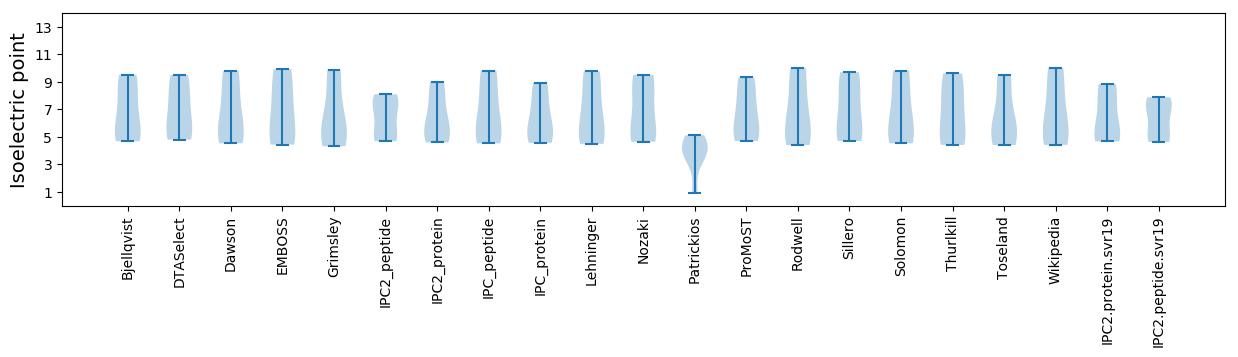
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A2D2ALL9|A0A2D2ALL9_9PAPI Replication protein E1 OS=Gammapapillomavirus 11 OX=1513256 GN=E1 PE=3 SV=1
MM1 pKa = 6.94NQSDD5 pKa = 3.78LAKK8 pKa = 10.6RR9 pKa = 11.84LDD11 pKa = 3.76ALQVQLMNLYY21 pKa = 10.29EE22 pKa = 4.62KK23 pKa = 11.03GPTDD27 pKa = 3.37LASQIRR33 pKa = 11.84HH34 pKa = 4.87FQLLRR39 pKa = 11.84KK40 pKa = 9.51EE41 pKa = 4.45SVLEE45 pKa = 3.79YY46 pKa = 9.75YY47 pKa = 10.17ARR49 pKa = 11.84KK50 pKa = 9.57EE51 pKa = 4.07GYY53 pKa = 9.42EE54 pKa = 3.8NLGLHH59 pKa = 6.55HH60 pKa = 7.19LPVLKK65 pKa = 10.76VSEE68 pKa = 4.69HH69 pKa = 5.67NAKK72 pKa = 9.86QAIKK76 pKa = 9.55MILYY80 pKa = 10.32LEE82 pKa = 4.48SLNKK86 pKa = 9.7SAYY89 pKa = 9.96ASEE92 pKa = 4.21EE93 pKa = 3.67WTLADD98 pKa = 3.56TSADD102 pKa = 3.53RR103 pKa = 11.84FLSPPKK109 pKa = 10.29NCFKK113 pKa = 10.66KK114 pKa = 10.41GSYY117 pKa = 8.36EE118 pKa = 4.06VEE120 pKa = 3.7VWFDD124 pKa = 3.73NDD126 pKa = 3.4PKK128 pKa = 11.22NAFPYY133 pKa = 10.51INWKK137 pKa = 8.35WIYY140 pKa = 10.11YY141 pKa = 9.34QDD143 pKa = 5.48NNEE146 pKa = 3.94QWHH149 pKa = 5.84KK150 pKa = 11.29VPGKK154 pKa = 8.57TDD156 pKa = 3.36YY157 pKa = 11.16NGLYY161 pKa = 10.01FVEE164 pKa = 4.99HH165 pKa = 7.64DD166 pKa = 3.5GTQTYY171 pKa = 10.37FLLFEE176 pKa = 5.24KK177 pKa = 10.61DD178 pKa = 2.95ANRR181 pKa = 11.84YY182 pKa = 8.43GNSGEE187 pKa = 4.09WTVNLPNEE195 pKa = 4.4QILPPSSTSSSRR207 pKa = 11.84RR208 pKa = 11.84HH209 pKa = 6.02LPDD212 pKa = 3.31SPEE215 pKa = 4.08VNRR218 pKa = 11.84GASTSYY224 pKa = 11.08SPTQEE229 pKa = 4.29EE230 pKa = 3.91IRR232 pKa = 11.84GRR234 pKa = 11.84QVQPPQTSPSTTTRR248 pKa = 11.84SPRR251 pKa = 11.84RR252 pKa = 11.84RR253 pKa = 11.84RR254 pKa = 11.84RR255 pKa = 11.84RR256 pKa = 11.84RR257 pKa = 11.84EE258 pKa = 3.98GEE260 pKa = 3.96SPPTKK265 pKa = 9.67RR266 pKa = 11.84RR267 pKa = 11.84RR268 pKa = 11.84GAQRR272 pKa = 11.84DD273 pKa = 4.06GGSSTISPEE282 pKa = 3.94AVGEE286 pKa = 4.14SHH288 pKa = 6.78QSVRR292 pKa = 11.84ATNLTRR298 pKa = 11.84VEE300 pKa = 4.01RR301 pKa = 11.84LKK303 pKa = 11.3AEE305 pKa = 4.15ARR307 pKa = 11.84DD308 pKa = 3.61PPLIIIRR315 pKa = 11.84GLANNLKK322 pKa = 8.85CWRR325 pKa = 11.84YY326 pKa = 10.01RR327 pKa = 11.84YY328 pKa = 7.49TKK330 pKa = 10.7KK331 pKa = 10.4NVKK334 pKa = 9.96NFIVMSSVFNWITNDD349 pKa = 3.11ANLDD353 pKa = 3.47GGRR356 pKa = 11.84ILVSFASVTQRR367 pKa = 11.84TQFLKK372 pKa = 10.9DD373 pKa = 3.26LTLPPGMSYY382 pKa = 11.23SLGNLEE388 pKa = 4.75KK389 pKa = 10.96LL390 pKa = 3.74
MM1 pKa = 6.94NQSDD5 pKa = 3.78LAKK8 pKa = 10.6RR9 pKa = 11.84LDD11 pKa = 3.76ALQVQLMNLYY21 pKa = 10.29EE22 pKa = 4.62KK23 pKa = 11.03GPTDD27 pKa = 3.37LASQIRR33 pKa = 11.84HH34 pKa = 4.87FQLLRR39 pKa = 11.84KK40 pKa = 9.51EE41 pKa = 4.45SVLEE45 pKa = 3.79YY46 pKa = 9.75YY47 pKa = 10.17ARR49 pKa = 11.84KK50 pKa = 9.57EE51 pKa = 4.07GYY53 pKa = 9.42EE54 pKa = 3.8NLGLHH59 pKa = 6.55HH60 pKa = 7.19LPVLKK65 pKa = 10.76VSEE68 pKa = 4.69HH69 pKa = 5.67NAKK72 pKa = 9.86QAIKK76 pKa = 9.55MILYY80 pKa = 10.32LEE82 pKa = 4.48SLNKK86 pKa = 9.7SAYY89 pKa = 9.96ASEE92 pKa = 4.21EE93 pKa = 3.67WTLADD98 pKa = 3.56TSADD102 pKa = 3.53RR103 pKa = 11.84FLSPPKK109 pKa = 10.29NCFKK113 pKa = 10.66KK114 pKa = 10.41GSYY117 pKa = 8.36EE118 pKa = 4.06VEE120 pKa = 3.7VWFDD124 pKa = 3.73NDD126 pKa = 3.4PKK128 pKa = 11.22NAFPYY133 pKa = 10.51INWKK137 pKa = 8.35WIYY140 pKa = 10.11YY141 pKa = 9.34QDD143 pKa = 5.48NNEE146 pKa = 3.94QWHH149 pKa = 5.84KK150 pKa = 11.29VPGKK154 pKa = 8.57TDD156 pKa = 3.36YY157 pKa = 11.16NGLYY161 pKa = 10.01FVEE164 pKa = 4.99HH165 pKa = 7.64DD166 pKa = 3.5GTQTYY171 pKa = 10.37FLLFEE176 pKa = 5.24KK177 pKa = 10.61DD178 pKa = 2.95ANRR181 pKa = 11.84YY182 pKa = 8.43GNSGEE187 pKa = 4.09WTVNLPNEE195 pKa = 4.4QILPPSSTSSSRR207 pKa = 11.84RR208 pKa = 11.84HH209 pKa = 6.02LPDD212 pKa = 3.31SPEE215 pKa = 4.08VNRR218 pKa = 11.84GASTSYY224 pKa = 11.08SPTQEE229 pKa = 4.29EE230 pKa = 3.91IRR232 pKa = 11.84GRR234 pKa = 11.84QVQPPQTSPSTTTRR248 pKa = 11.84SPRR251 pKa = 11.84RR252 pKa = 11.84RR253 pKa = 11.84RR254 pKa = 11.84RR255 pKa = 11.84RR256 pKa = 11.84RR257 pKa = 11.84EE258 pKa = 3.98GEE260 pKa = 3.96SPPTKK265 pKa = 9.67RR266 pKa = 11.84RR267 pKa = 11.84RR268 pKa = 11.84GAQRR272 pKa = 11.84DD273 pKa = 4.06GGSSTISPEE282 pKa = 3.94AVGEE286 pKa = 4.14SHH288 pKa = 6.78QSVRR292 pKa = 11.84ATNLTRR298 pKa = 11.84VEE300 pKa = 4.01RR301 pKa = 11.84LKK303 pKa = 11.3AEE305 pKa = 4.15ARR307 pKa = 11.84DD308 pKa = 3.61PPLIIIRR315 pKa = 11.84GLANNLKK322 pKa = 8.85CWRR325 pKa = 11.84YY326 pKa = 10.01RR327 pKa = 11.84YY328 pKa = 7.49TKK330 pKa = 10.7KK331 pKa = 10.4NVKK334 pKa = 9.96NFIVMSSVFNWITNDD349 pKa = 3.11ANLDD353 pKa = 3.47GGRR356 pKa = 11.84ILVSFASVTQRR367 pKa = 11.84TQFLKK372 pKa = 10.9DD373 pKa = 3.26LTLPPGMSYY382 pKa = 11.23SLGNLEE388 pKa = 4.75KK389 pKa = 10.96LL390 pKa = 3.74
Molecular weight: 44.88 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
2483 |
98 |
603 |
354.7 |
40.4 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
5.518 ± 0.378 | 2.296 ± 0.799 |
6.242 ± 0.399 | 6.122 ± 0.318 |
4.752 ± 0.521 | 5.034 ± 0.689 |
1.973 ± 0.334 | 5.961 ± 0.884 |
5.679 ± 0.804 | 9.344 ± 1.226 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.135 ± 0.457 | 5.558 ± 0.571 |
6.122 ± 0.81 | 4.631 ± 0.528 |
5.598 ± 0.716 | 6.726 ± 0.767 |
5.92 ± 0.555 | 5.558 ± 0.497 |
1.248 ± 0.272 | 3.584 ± 0.422 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
