
Drosophila sturtevanti sigmavirus
Taxonomy: Viruses; Riboviria; Orthornavirae; Negarnaviricota; Haploviricotina; Monjiviricetes; Mononegavirales; Rhabdoviridae; Alpharhabdovirinae; Sigmavirus; Sturtevanti sigmavirus
Average proteome isoelectric point is 6.34
Get precalculated fractions of proteins
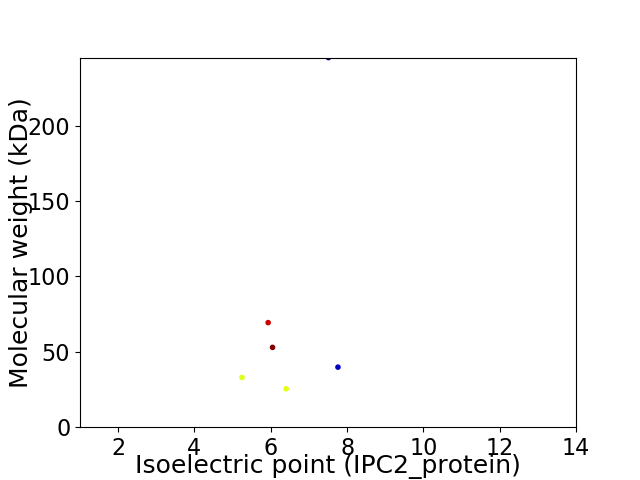
Virtual 2D-PAGE plot for 6 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A140D8M6|A0A140D8M6_9RHAB PP3 OS=Drosophila sturtevanti sigmavirus OX=1802946 GN=X PE=4 SV=1
MM1 pKa = 7.62ANCKK5 pKa = 8.57PTFEE9 pKa = 4.87LSHH12 pKa = 6.3EE13 pKa = 4.68LDD15 pKa = 5.05HH16 pKa = 7.56IWATTRR22 pKa = 11.84KK23 pKa = 9.58AIKK26 pKa = 10.0ALPKK30 pKa = 10.3LDD32 pKa = 4.07TNLKK36 pKa = 9.96SDD38 pKa = 3.36QNYY41 pKa = 10.14GDD43 pKa = 3.63NHH45 pKa = 6.67PDD47 pKa = 3.06KK48 pKa = 9.4TTLQSKK54 pKa = 8.18ITQQNSSGEE63 pKa = 4.13SDD65 pKa = 3.11SWAVLAMEE73 pKa = 4.99EE74 pKa = 4.49PDD76 pKa = 3.64NKK78 pKa = 10.67NISAEE83 pKa = 4.11VEE85 pKa = 3.72PDD87 pKa = 2.92QWPDD91 pKa = 3.17FDD93 pKa = 6.66DD94 pKa = 5.66SDD96 pKa = 3.99DD97 pKa = 3.79TSQEE101 pKa = 4.02LEE103 pKa = 4.05EE104 pKa = 6.18DD105 pKa = 3.48EE106 pKa = 5.11LFPIFQMKK114 pKa = 9.25NDD116 pKa = 3.29KK117 pKa = 9.82RR118 pKa = 11.84AEE120 pKa = 4.1MFSVPEE126 pKa = 4.02VEE128 pKa = 5.6QMVKK132 pKa = 10.88DD133 pKa = 4.3LLYY136 pKa = 10.7QLNLSNHH143 pKa = 6.94DD144 pKa = 3.53YY145 pKa = 10.61SYY147 pKa = 10.7HH148 pKa = 6.61IEE150 pKa = 4.04GTIRR154 pKa = 11.84RR155 pKa = 11.84PVLHH159 pKa = 5.82VNKK162 pKa = 10.04RR163 pKa = 11.84LTSPTSNVQPSTSKK177 pKa = 10.83RR178 pKa = 11.84IQTPEE183 pKa = 3.99NKK185 pKa = 9.24TKK187 pKa = 9.68STPPKK192 pKa = 10.58AVDD195 pKa = 3.29QEE197 pKa = 4.46SFNAVDD203 pKa = 3.57HH204 pKa = 6.97RR205 pKa = 11.84YY206 pKa = 10.12NDD208 pKa = 3.92LMRR211 pKa = 11.84KK212 pKa = 9.55NYY214 pKa = 7.93TLNYY218 pKa = 8.46IDD220 pKa = 4.0STIKK224 pKa = 10.27SKK226 pKa = 11.26YY227 pKa = 9.73VINKK231 pKa = 7.48EE232 pKa = 3.81NPDD235 pKa = 3.43FKK237 pKa = 10.76KK238 pKa = 10.51SYY240 pKa = 9.73FVEE243 pKa = 4.67VYY245 pKa = 9.93GANGKK250 pKa = 9.59FPALDD255 pKa = 2.8IHH257 pKa = 7.27LIQNILTIQKK267 pKa = 9.44KK268 pKa = 4.5WHH270 pKa = 5.5QMLNLVDD277 pKa = 4.38FTPTYY282 pKa = 7.53TTVV285 pKa = 2.66
MM1 pKa = 7.62ANCKK5 pKa = 8.57PTFEE9 pKa = 4.87LSHH12 pKa = 6.3EE13 pKa = 4.68LDD15 pKa = 5.05HH16 pKa = 7.56IWATTRR22 pKa = 11.84KK23 pKa = 9.58AIKK26 pKa = 10.0ALPKK30 pKa = 10.3LDD32 pKa = 4.07TNLKK36 pKa = 9.96SDD38 pKa = 3.36QNYY41 pKa = 10.14GDD43 pKa = 3.63NHH45 pKa = 6.67PDD47 pKa = 3.06KK48 pKa = 9.4TTLQSKK54 pKa = 8.18ITQQNSSGEE63 pKa = 4.13SDD65 pKa = 3.11SWAVLAMEE73 pKa = 4.99EE74 pKa = 4.49PDD76 pKa = 3.64NKK78 pKa = 10.67NISAEE83 pKa = 4.11VEE85 pKa = 3.72PDD87 pKa = 2.92QWPDD91 pKa = 3.17FDD93 pKa = 6.66DD94 pKa = 5.66SDD96 pKa = 3.99DD97 pKa = 3.79TSQEE101 pKa = 4.02LEE103 pKa = 4.05EE104 pKa = 6.18DD105 pKa = 3.48EE106 pKa = 5.11LFPIFQMKK114 pKa = 9.25NDD116 pKa = 3.29KK117 pKa = 9.82RR118 pKa = 11.84AEE120 pKa = 4.1MFSVPEE126 pKa = 4.02VEE128 pKa = 5.6QMVKK132 pKa = 10.88DD133 pKa = 4.3LLYY136 pKa = 10.7QLNLSNHH143 pKa = 6.94DD144 pKa = 3.53YY145 pKa = 10.61SYY147 pKa = 10.7HH148 pKa = 6.61IEE150 pKa = 4.04GTIRR154 pKa = 11.84RR155 pKa = 11.84PVLHH159 pKa = 5.82VNKK162 pKa = 10.04RR163 pKa = 11.84LTSPTSNVQPSTSKK177 pKa = 10.83RR178 pKa = 11.84IQTPEE183 pKa = 3.99NKK185 pKa = 9.24TKK187 pKa = 9.68STPPKK192 pKa = 10.58AVDD195 pKa = 3.29QEE197 pKa = 4.46SFNAVDD203 pKa = 3.57HH204 pKa = 6.97RR205 pKa = 11.84YY206 pKa = 10.12NDD208 pKa = 3.92LMRR211 pKa = 11.84KK212 pKa = 9.55NYY214 pKa = 7.93TLNYY218 pKa = 8.46IDD220 pKa = 4.0STIKK224 pKa = 10.27SKK226 pKa = 11.26YY227 pKa = 9.73VINKK231 pKa = 7.48EE232 pKa = 3.81NPDD235 pKa = 3.43FKK237 pKa = 10.76KK238 pKa = 10.51SYY240 pKa = 9.73FVEE243 pKa = 4.67VYY245 pKa = 9.93GANGKK250 pKa = 9.59FPALDD255 pKa = 2.8IHH257 pKa = 7.27LIQNILTIQKK267 pKa = 9.44KK268 pKa = 4.5WHH270 pKa = 5.5QMLNLVDD277 pKa = 4.38FTPTYY282 pKa = 7.53TTVV285 pKa = 2.66
Molecular weight: 32.99 kDa
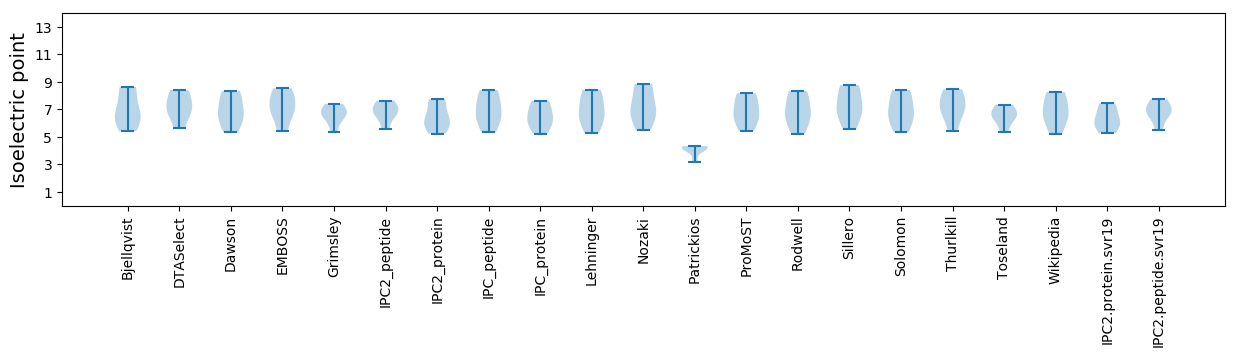
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A140D8M7|A0A140D8M7_9RHAB Matrix protein OS=Drosophila sturtevanti sigmavirus OX=1802946 GN=M PE=4 SV=1
MM1 pKa = 7.36SSSHH5 pKa = 6.77RR6 pKa = 11.84KK7 pKa = 6.62TSTYY11 pKa = 10.59IMKK14 pKa = 8.91KK15 pKa = 5.12TTLIVVVLSVVQVSQLTNLTCRR37 pKa = 11.84CEE39 pKa = 4.72LSDD42 pKa = 5.0LTDD45 pKa = 3.32STNDD49 pKa = 3.07ILVANVTYY57 pKa = 10.33DD58 pKa = 3.58LPHH61 pKa = 6.87NDD63 pKa = 4.74PYY65 pKa = 11.42NISQSINVNYY75 pKa = 10.01WNGTEE80 pKa = 4.04LVKK83 pKa = 10.12TKK85 pKa = 10.02QHH87 pKa = 5.73LCVYY91 pKa = 8.85LTDD94 pKa = 3.81EE95 pKa = 4.52SGMTVYY101 pKa = 10.96SNPDD105 pKa = 3.19DD106 pKa = 3.53TTKK109 pKa = 10.82NVEE112 pKa = 4.11SVIRR116 pKa = 11.84EE117 pKa = 3.77IKK119 pKa = 8.65TNEE122 pKa = 4.08GVPHH126 pKa = 7.66DD127 pKa = 5.55DD128 pKa = 3.44LTQSEE133 pKa = 5.05LLCTMGMCRR142 pKa = 11.84DD143 pKa = 3.41RR144 pKa = 11.84KK145 pKa = 10.69GSFKK149 pKa = 10.32EE150 pKa = 3.72ANRR153 pKa = 11.84GIRR156 pKa = 11.84NLNSILTLKK165 pKa = 10.9VNLTKK170 pKa = 10.79AGVPNEE176 pKa = 4.03RR177 pKa = 11.84SLMDD181 pKa = 3.87KK182 pKa = 10.84VNQQLMTSNHH192 pKa = 5.96IPNMGHH198 pKa = 6.65SSLLAQDD205 pKa = 4.34YY206 pKa = 10.55LFSRR210 pKa = 11.84PDD212 pKa = 3.05EE213 pKa = 4.38RR214 pKa = 11.84MEE216 pKa = 6.49DD217 pKa = 2.96IMFPSGEE224 pKa = 3.86HH225 pKa = 5.1RR226 pKa = 11.84RR227 pKa = 11.84RR228 pKa = 11.84RR229 pKa = 11.84SLEE232 pKa = 3.87SEE234 pKa = 3.87TSRR237 pKa = 11.84EE238 pKa = 3.36IRR240 pKa = 11.84AWRR243 pKa = 11.84IRR245 pKa = 11.84RR246 pKa = 11.84LIKK249 pKa = 10.45KK250 pKa = 9.63IVGRR254 pKa = 11.84LEE256 pKa = 3.99KK257 pKa = 11.14YY258 pKa = 9.69EE259 pKa = 4.14VMWLEE264 pKa = 4.18NYY266 pKa = 9.13TLFLVYY272 pKa = 10.46LRR274 pKa = 11.84QSPHH278 pKa = 6.46FDD280 pKa = 2.82SSACVLHH287 pKa = 6.57PLFSDD292 pKa = 4.31LLKK295 pKa = 10.76CYY297 pKa = 10.07SANLHH302 pKa = 5.5VMNNTKK308 pKa = 10.46HH309 pKa = 6.3GVDD312 pKa = 3.08AMKK315 pKa = 10.89YY316 pKa = 8.71FAEE319 pKa = 4.65CVHH322 pKa = 6.49IKK324 pKa = 10.31YY325 pKa = 9.24SHH327 pKa = 6.74KK328 pKa = 10.12MEE330 pKa = 4.01QFMKK334 pKa = 10.53NYY336 pKa = 9.69LRR338 pKa = 11.84RR339 pKa = 11.84DD340 pKa = 3.62LLSLL344 pKa = 3.83
MM1 pKa = 7.36SSSHH5 pKa = 6.77RR6 pKa = 11.84KK7 pKa = 6.62TSTYY11 pKa = 10.59IMKK14 pKa = 8.91KK15 pKa = 5.12TTLIVVVLSVVQVSQLTNLTCRR37 pKa = 11.84CEE39 pKa = 4.72LSDD42 pKa = 5.0LTDD45 pKa = 3.32STNDD49 pKa = 3.07ILVANVTYY57 pKa = 10.33DD58 pKa = 3.58LPHH61 pKa = 6.87NDD63 pKa = 4.74PYY65 pKa = 11.42NISQSINVNYY75 pKa = 10.01WNGTEE80 pKa = 4.04LVKK83 pKa = 10.12TKK85 pKa = 10.02QHH87 pKa = 5.73LCVYY91 pKa = 8.85LTDD94 pKa = 3.81EE95 pKa = 4.52SGMTVYY101 pKa = 10.96SNPDD105 pKa = 3.19DD106 pKa = 3.53TTKK109 pKa = 10.82NVEE112 pKa = 4.11SVIRR116 pKa = 11.84EE117 pKa = 3.77IKK119 pKa = 8.65TNEE122 pKa = 4.08GVPHH126 pKa = 7.66DD127 pKa = 5.55DD128 pKa = 3.44LTQSEE133 pKa = 5.05LLCTMGMCRR142 pKa = 11.84DD143 pKa = 3.41RR144 pKa = 11.84KK145 pKa = 10.69GSFKK149 pKa = 10.32EE150 pKa = 3.72ANRR153 pKa = 11.84GIRR156 pKa = 11.84NLNSILTLKK165 pKa = 10.9VNLTKK170 pKa = 10.79AGVPNEE176 pKa = 4.03RR177 pKa = 11.84SLMDD181 pKa = 3.87KK182 pKa = 10.84VNQQLMTSNHH192 pKa = 5.96IPNMGHH198 pKa = 6.65SSLLAQDD205 pKa = 4.34YY206 pKa = 10.55LFSRR210 pKa = 11.84PDD212 pKa = 3.05EE213 pKa = 4.38RR214 pKa = 11.84MEE216 pKa = 6.49DD217 pKa = 2.96IMFPSGEE224 pKa = 3.86HH225 pKa = 5.1RR226 pKa = 11.84RR227 pKa = 11.84RR228 pKa = 11.84RR229 pKa = 11.84SLEE232 pKa = 3.87SEE234 pKa = 3.87TSRR237 pKa = 11.84EE238 pKa = 3.36IRR240 pKa = 11.84AWRR243 pKa = 11.84IRR245 pKa = 11.84RR246 pKa = 11.84LIKK249 pKa = 10.45KK250 pKa = 9.63IVGRR254 pKa = 11.84LEE256 pKa = 3.99KK257 pKa = 11.14YY258 pKa = 9.69EE259 pKa = 4.14VMWLEE264 pKa = 4.18NYY266 pKa = 9.13TLFLVYY272 pKa = 10.46LRR274 pKa = 11.84QSPHH278 pKa = 6.46FDD280 pKa = 2.82SSACVLHH287 pKa = 6.57PLFSDD292 pKa = 4.31LLKK295 pKa = 10.76CYY297 pKa = 10.07SANLHH302 pKa = 5.5VMNNTKK308 pKa = 10.46HH309 pKa = 6.3GVDD312 pKa = 3.08AMKK315 pKa = 10.89YY316 pKa = 8.71FAEE319 pKa = 4.65CVHH322 pKa = 6.49IKK324 pKa = 10.31YY325 pKa = 9.24SHH327 pKa = 6.74KK328 pKa = 10.12MEE330 pKa = 4.01QFMKK334 pKa = 10.53NYY336 pKa = 9.69LRR338 pKa = 11.84RR339 pKa = 11.84DD340 pKa = 3.62LLSLL344 pKa = 3.83
Molecular weight: 39.84 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
4082 |
224 |
2144 |
680.3 |
77.65 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
4.361 ± 0.492 | 1.886 ± 0.268 |
5.585 ± 0.348 | 5.537 ± 0.259 |
4.312 ± 0.392 | 5.855 ± 0.748 |
2.964 ± 0.198 | 6.786 ± 0.474 |
5.928 ± 0.555 | 10.51 ± 0.609 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.401 ± 0.287 | 4.679 ± 0.538 |
5.341 ± 0.382 | 3.577 ± 0.218 |
5.169 ± 0.54 | 7.545 ± 0.588 |
5.781 ± 0.442 | 5.904 ± 0.433 |
1.788 ± 0.203 | 4.067 ± 0.323 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
