
Mona Grita virus
Taxonomy: Viruses; Riboviria; Orthornavirae; Negarnaviricota; Polyploviricotina; Ellioviricetes; Bunyavirales; Phenuiviridae; Phlebovirus; Mona Grita phlebovirus
Average proteome isoelectric point is 6.43
Get precalculated fractions of proteins
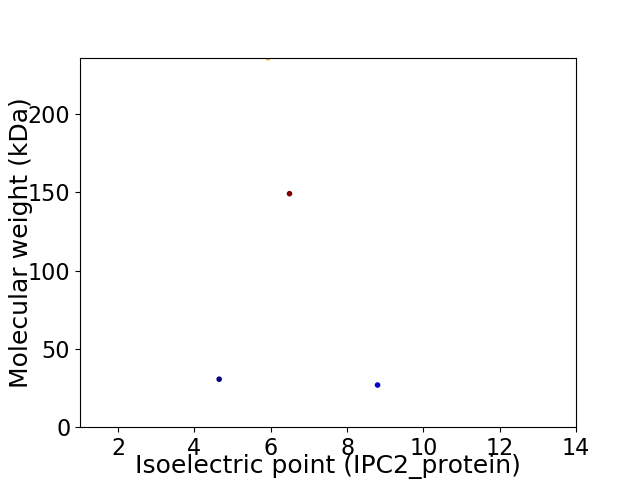
Virtual 2D-PAGE plot for 4 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A482KEM8|A0A482KEM8_9VIRU Nonstructural protein OS=Mona Grita virus OX=2559111 GN=NSs PE=4 SV=1
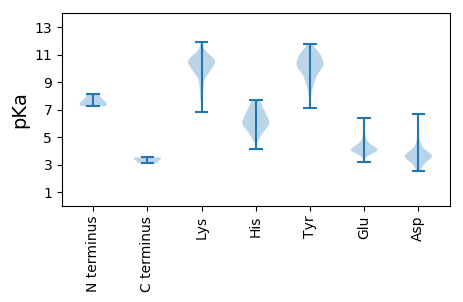
MM1 pKa = 7.36NFLYY5 pKa = 10.04DD6 pKa = 3.34QPVIFRR12 pKa = 11.84MGGVGDD18 pKa = 4.12PEE20 pKa = 4.55VEE22 pKa = 3.97VGYY25 pKa = 10.77LPFEE29 pKa = 4.11AWSVRR34 pKa = 11.84KK35 pKa = 9.71ICVHH39 pKa = 5.11NTMEE43 pKa = 4.95IPLEE47 pKa = 4.19TYY49 pKa = 10.13RR50 pKa = 11.84FAMDD54 pKa = 3.55HH55 pKa = 6.68RR56 pKa = 11.84PTLRR60 pKa = 11.84DD61 pKa = 3.82FYY63 pKa = 11.53SRR65 pKa = 11.84GEE67 pKa = 3.99FPFRR71 pKa = 11.84WGPGTFNSRR80 pKa = 11.84VQEE83 pKa = 4.02EE84 pKa = 4.73TTNSFDD90 pKa = 3.68SAIMDD95 pKa = 3.83LVGFPLASFTKK106 pKa = 10.53SFLPNVKK113 pKa = 9.96LALSWPLGYY122 pKa = 7.71PTTDD126 pKa = 4.54FIQLSGIPGHH136 pKa = 5.71FTHH139 pKa = 6.76NGWKK143 pKa = 10.31GMAATSLFRR152 pKa = 11.84MNPQLEE158 pKa = 4.32RR159 pKa = 11.84FDD161 pKa = 3.76QVFVEE166 pKa = 3.99AHH168 pKa = 5.69KK169 pKa = 11.07SILEE173 pKa = 3.86EE174 pKa = 3.99AEE176 pKa = 3.99RR177 pKa = 11.84RR178 pKa = 11.84GLSSDD183 pKa = 3.52YY184 pKa = 10.08FTGYY188 pKa = 11.12DD189 pKa = 3.61LLKK192 pKa = 10.51EE193 pKa = 4.27VASLQIVRR201 pKa = 11.84LLNAVDD207 pKa = 3.34IDD209 pKa = 4.06VVYY212 pKa = 11.38SNIPTNLTYY221 pKa = 11.18KK222 pKa = 10.9LLDD225 pKa = 3.72LRR227 pKa = 11.84DD228 pKa = 3.76TFEE231 pKa = 4.4NEE233 pKa = 4.05SPEE236 pKa = 4.09LLGNRR241 pKa = 11.84FWIPSPDD248 pKa = 2.81TWFDD252 pKa = 3.35GLYY255 pKa = 10.3LQTDD259 pKa = 3.67SSFSSDD265 pKa = 2.77EE266 pKa = 4.09DD267 pKa = 3.51
MM1 pKa = 7.36NFLYY5 pKa = 10.04DD6 pKa = 3.34QPVIFRR12 pKa = 11.84MGGVGDD18 pKa = 4.12PEE20 pKa = 4.55VEE22 pKa = 3.97VGYY25 pKa = 10.77LPFEE29 pKa = 4.11AWSVRR34 pKa = 11.84KK35 pKa = 9.71ICVHH39 pKa = 5.11NTMEE43 pKa = 4.95IPLEE47 pKa = 4.19TYY49 pKa = 10.13RR50 pKa = 11.84FAMDD54 pKa = 3.55HH55 pKa = 6.68RR56 pKa = 11.84PTLRR60 pKa = 11.84DD61 pKa = 3.82FYY63 pKa = 11.53SRR65 pKa = 11.84GEE67 pKa = 3.99FPFRR71 pKa = 11.84WGPGTFNSRR80 pKa = 11.84VQEE83 pKa = 4.02EE84 pKa = 4.73TTNSFDD90 pKa = 3.68SAIMDD95 pKa = 3.83LVGFPLASFTKK106 pKa = 10.53SFLPNVKK113 pKa = 9.96LALSWPLGYY122 pKa = 7.71PTTDD126 pKa = 4.54FIQLSGIPGHH136 pKa = 5.71FTHH139 pKa = 6.76NGWKK143 pKa = 10.31GMAATSLFRR152 pKa = 11.84MNPQLEE158 pKa = 4.32RR159 pKa = 11.84FDD161 pKa = 3.76QVFVEE166 pKa = 3.99AHH168 pKa = 5.69KK169 pKa = 11.07SILEE173 pKa = 3.86EE174 pKa = 3.99AEE176 pKa = 3.99RR177 pKa = 11.84RR178 pKa = 11.84GLSSDD183 pKa = 3.52YY184 pKa = 10.08FTGYY188 pKa = 11.12DD189 pKa = 3.61LLKK192 pKa = 10.51EE193 pKa = 4.27VASLQIVRR201 pKa = 11.84LLNAVDD207 pKa = 3.34IDD209 pKa = 4.06VVYY212 pKa = 11.38SNIPTNLTYY221 pKa = 11.18KK222 pKa = 10.9LLDD225 pKa = 3.72LRR227 pKa = 11.84DD228 pKa = 3.76TFEE231 pKa = 4.4NEE233 pKa = 4.05SPEE236 pKa = 4.09LLGNRR241 pKa = 11.84FWIPSPDD248 pKa = 2.81TWFDD252 pKa = 3.35GLYY255 pKa = 10.3LQTDD259 pKa = 3.67SSFSSDD265 pKa = 2.77EE266 pKa = 4.09DD267 pKa = 3.51
Molecular weight: 30.68 kDa
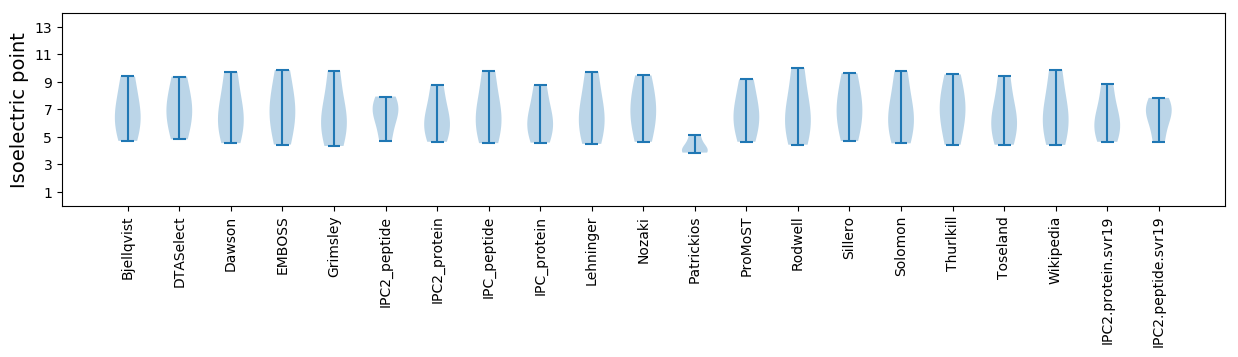
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A482KC00|A0A482KC00_9VIRU Glycoprotein OS=Mona Grita virus OX=2559111 GN=GPC PE=4 SV=1
MM1 pKa = 7.56TDD3 pKa = 3.19YY4 pKa = 11.74AEE6 pKa = 4.07IAIAFAGEE14 pKa = 4.34PINNAEE20 pKa = 3.85VMGWVNEE27 pKa = 3.96FAYY30 pKa = 10.14EE31 pKa = 4.14GFSAQRR37 pKa = 11.84IIQLVQEE44 pKa = 5.25RR45 pKa = 11.84GPQTWQVDD53 pKa = 3.9VKK55 pKa = 10.81MMIVLALTRR64 pKa = 11.84GNKK67 pKa = 7.49PSKK70 pKa = 10.02MIEE73 pKa = 3.94KK74 pKa = 9.86MSAEE78 pKa = 4.22GKK80 pKa = 10.19KK81 pKa = 10.14KK82 pKa = 9.12ATRR85 pKa = 11.84LITTYY90 pKa = 10.48NLKK93 pKa = 10.41SGNPGRR99 pKa = 11.84DD100 pKa = 3.45DD101 pKa = 3.54LTLSRR106 pKa = 11.84VASAFAGWTCQALAVLHH123 pKa = 6.86PYY125 pKa = 10.72LPVTGASMDD134 pKa = 4.42SISPNYY140 pKa = 8.29PRR142 pKa = 11.84AMMHH146 pKa = 6.7PSFAGLIDD154 pKa = 3.52NSIPEE159 pKa = 4.71AYY161 pKa = 9.95LQLVVDD167 pKa = 3.84AHH169 pKa = 6.94ALYY172 pKa = 10.84LLQFSRR178 pKa = 11.84VINKK182 pKa = 8.82NMRR185 pKa = 11.84GQPKK189 pKa = 9.72SVVVSSFLQPMNAAIVSGFISNDD212 pKa = 2.71KK213 pKa = 9.22RR214 pKa = 11.84RR215 pKa = 11.84KK216 pKa = 8.2MLMAFGIVDD225 pKa = 3.8QNGKK229 pKa = 7.34PTQAVEE235 pKa = 4.17TAAKK239 pKa = 10.82AFMTINN245 pKa = 3.28
MM1 pKa = 7.56TDD3 pKa = 3.19YY4 pKa = 11.74AEE6 pKa = 4.07IAIAFAGEE14 pKa = 4.34PINNAEE20 pKa = 3.85VMGWVNEE27 pKa = 3.96FAYY30 pKa = 10.14EE31 pKa = 4.14GFSAQRR37 pKa = 11.84IIQLVQEE44 pKa = 5.25RR45 pKa = 11.84GPQTWQVDD53 pKa = 3.9VKK55 pKa = 10.81MMIVLALTRR64 pKa = 11.84GNKK67 pKa = 7.49PSKK70 pKa = 10.02MIEE73 pKa = 3.94KK74 pKa = 9.86MSAEE78 pKa = 4.22GKK80 pKa = 10.19KK81 pKa = 10.14KK82 pKa = 9.12ATRR85 pKa = 11.84LITTYY90 pKa = 10.48NLKK93 pKa = 10.41SGNPGRR99 pKa = 11.84DD100 pKa = 3.45DD101 pKa = 3.54LTLSRR106 pKa = 11.84VASAFAGWTCQALAVLHH123 pKa = 6.86PYY125 pKa = 10.72LPVTGASMDD134 pKa = 4.42SISPNYY140 pKa = 8.29PRR142 pKa = 11.84AMMHH146 pKa = 6.7PSFAGLIDD154 pKa = 3.52NSIPEE159 pKa = 4.71AYY161 pKa = 9.95LQLVVDD167 pKa = 3.84AHH169 pKa = 6.94ALYY172 pKa = 10.84LLQFSRR178 pKa = 11.84VINKK182 pKa = 8.82NMRR185 pKa = 11.84GQPKK189 pKa = 9.72SVVVSSFLQPMNAAIVSGFISNDD212 pKa = 2.71KK213 pKa = 9.22RR214 pKa = 11.84RR215 pKa = 11.84KK216 pKa = 8.2MLMAFGIVDD225 pKa = 3.8QNGKK229 pKa = 7.34PTQAVEE235 pKa = 4.17TAAKK239 pKa = 10.82AFMTINN245 pKa = 3.28
Molecular weight: 26.95 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
3947 |
245 |
2083 |
986.8 |
110.76 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
4.636 ± 1.085 | 2.584 ± 0.837 |
5.194 ± 0.562 | 6.587 ± 0.461 |
4.864 ± 0.573 | 6.486 ± 0.257 |
2.128 ± 0.176 | 6.714 ± 0.42 |
6.283 ± 0.659 | 9.121 ± 0.47 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
3.116 ± 0.408 | 4.18 ± 0.487 |
4.054 ± 0.478 | 3.344 ± 0.216 |
4.814 ± 0.278 | 10.312 ± 0.823 |
5.194 ± 0.191 | 6.309 ± 0.199 |
1.267 ± 0.165 | 2.812 ± 0.16 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
