
Sphingomonas sp. HMWF008
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Alphaproteobacteria; Sphingomonadales; Sphingomonadaceae; Sphingomonas; unclassified Sphingomonas
Average proteome isoelectric point is 6.69
Get precalculated fractions of proteins
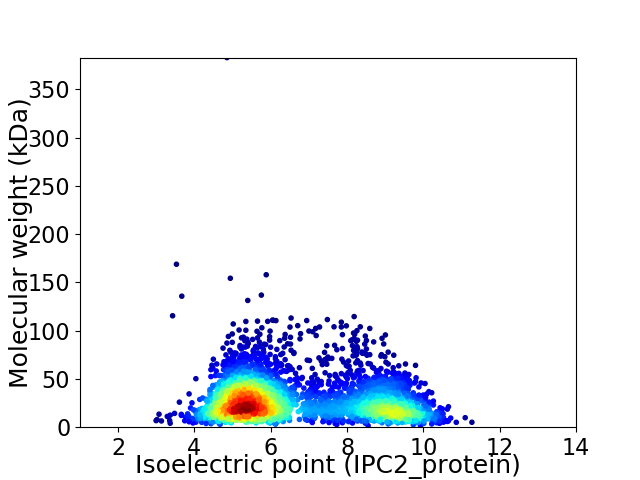
Virtual 2D-PAGE plot for 3537 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A2R7IN65|A0A2R7IN65_9SPHN UbiA family prenyltransferase OS=Sphingomonas sp. HMWF008 OX=2056845 GN=DBR17_17125 PE=4 SV=1
SS1 pKa = 4.87YY2 pKa = 9.85TFSVVVNGDD11 pKa = 3.47ALVEE15 pKa = 4.26PNEE18 pKa = 4.29TVLVNLSNVVGAAVTDD34 pKa = 3.98SQGSGTIIDD43 pKa = 3.92NDD45 pKa = 3.88FPTLSISDD53 pKa = 3.8ASVSEE58 pKa = 4.35GDD60 pKa = 3.35SGTTVLTYY68 pKa = 10.13TVTASAPALAGGISFTIATADD89 pKa = 3.48GTATAGSDD97 pKa = 3.29YY98 pKa = 10.83DD99 pKa = 3.86AQSTTGTIAEE109 pKa = 4.4GQTTYY114 pKa = 10.66TFSVVVNGDD123 pKa = 3.13VSNEE127 pKa = 4.0SNEE130 pKa = 4.24TVLVNLSNVTGATLVDD146 pKa = 3.68AQGVGTIVNDD156 pKa = 4.17DD157 pKa = 3.49GPIRR161 pKa = 11.84IFSEE165 pKa = 4.14SFSNFLAAGFAPTPSASQLDD185 pKa = 3.71SDD187 pKa = 3.82IWRR190 pKa = 11.84VAGLSDD196 pKa = 4.13NLTPAYY202 pKa = 10.0GATLTTGDD210 pKa = 3.88FARR213 pKa = 11.84GVINGAADD221 pKa = 3.59PTSGGVYY228 pKa = 10.14SPSANRR234 pKa = 11.84ALVVQPTGSDD244 pKa = 3.19FDD246 pKa = 3.98AGGFIEE252 pKa = 5.95AKK254 pKa = 9.43IQNSSGKK261 pKa = 7.61TAVAFDD267 pKa = 3.92VGFDD271 pKa = 3.0WAYY274 pKa = 10.6RR275 pKa = 11.84NSGNRR280 pKa = 11.84ADD282 pKa = 4.92NMQFSYY288 pKa = 9.61STDD291 pKa = 2.97GTNFTVVPAAAFVTPAAADD310 pKa = 3.47ASVAATFSNQNEE322 pKa = 4.61TVSLTGLAVAAGGFLYY338 pKa = 10.68LRR340 pKa = 11.84WTHH343 pKa = 6.86LDD345 pKa = 3.21GATGSGNRR353 pKa = 11.84DD354 pKa = 3.33EE355 pKa = 5.49IGIDD359 pKa = 3.23NVTIDD364 pKa = 3.64ATTSNLATATVANISLAEE382 pKa = 4.24GDD384 pKa = 4.3AGTTSATFTVVRR396 pKa = 11.84SSGVGAASIDD406 pKa = 3.73YY407 pKa = 7.65TTANGSAIAGTDD419 pKa = 3.46YY420 pKa = 10.96VAASGTVSFADD431 pKa = 3.92GEE433 pKa = 4.53TSQTVTILITGDD445 pKa = 3.33TAFEE449 pKa = 4.18AGEE452 pKa = 4.35TFFLNLSNPSNVLLPSSQASVTITNDD478 pKa = 3.02DD479 pKa = 3.66NGAVAIYY486 pKa = 9.47DD487 pKa = 3.55IQGAGHH493 pKa = 6.58TSALVGQFIQTSGIVTAVAANGFYY517 pKa = 10.83LQDD520 pKa = 3.49PTGDD524 pKa = 3.5GNTRR528 pKa = 11.84TSDD531 pKa = 3.75GIFVFTSTAPTVVVGNGISLTGTVTEE557 pKa = 5.06FDD559 pKa = 3.91AGLDD563 pKa = 3.78TLTVTQITAPTITVLTQTNTLPNAVLIGATGGILPPTQSIDD604 pKa = 3.3SDD606 pKa = 3.66NFTVFNPEE614 pKa = 2.83VDD616 pKa = 4.59GIDD619 pKa = 3.57FYY621 pKa = 11.46EE622 pKa = 4.26TLEE625 pKa = 4.08GMRR628 pKa = 11.84VTVQAPTVTSNTNSFGEE645 pKa = 4.32TFIVASGGIGATGVNEE661 pKa = 4.06RR662 pKa = 11.84GGITISAGDD671 pKa = 3.91NNPEE675 pKa = 4.77RR676 pKa = 11.84IQIDD680 pKa = 3.39NDD682 pKa = 3.63TTIFAGYY689 pKa = 7.75TPSHH693 pKa = 5.72TVGDD697 pKa = 3.81RR698 pKa = 11.84LGDD701 pKa = 3.4VTGVFNYY708 pKa = 10.25AFQSYY713 pKa = 8.46EE714 pKa = 4.28LIVTEE719 pKa = 4.37TVVVTNDD726 pKa = 3.26TTPTRR731 pKa = 11.84EE732 pKa = 4.04TTTLAGDD739 pKa = 3.62ATHH742 pKa = 5.97VTFASFNIEE751 pKa = 4.04NADD754 pKa = 3.6PTDD757 pKa = 3.97PASKK761 pKa = 8.98FTLLGTEE768 pKa = 3.57IRR770 pKa = 11.84YY771 pKa = 9.35ALNTPDD777 pKa = 4.83IIALQEE783 pKa = 3.93VQDD786 pKa = 4.09ADD788 pKa = 3.93GAGTGTNYY796 pKa = 10.61SGTVTAQKK804 pKa = 10.71LIDD807 pKa = 4.78AIIAAGGPTYY817 pKa = 10.61SYY819 pKa = 11.28IEE821 pKa = 4.08IAPTANNTTGGEE833 pKa = 4.06PNGNIRR839 pKa = 11.84NGFLYY844 pKa = 10.2NAARR848 pKa = 11.84VTYY851 pKa = 10.35VDD853 pKa = 3.54GSVALVPGIAYY864 pKa = 10.04NNSRR868 pKa = 11.84NPLVADD874 pKa = 4.74FSFNGQVFTTVSVHH888 pKa = 4.12STSRR892 pKa = 11.84GGSDD896 pKa = 3.47AFYY899 pKa = 11.29GDD901 pKa = 3.42IQPPVQAGDD910 pKa = 3.75SQRR913 pKa = 11.84TAQADD918 pKa = 3.69TLRR921 pKa = 11.84AYY923 pKa = 9.89INDD926 pKa = 3.69QLATDD931 pKa = 4.82PNHH934 pKa = 5.82QFIIGGDD941 pKa = 3.61FNGYY945 pKa = 9.52YY946 pKa = 9.9YY947 pKa = 8.81EE948 pKa = 4.1TAMQRR953 pKa = 11.84LVAGGVLTNLYY964 pKa = 10.5DD965 pKa = 4.21LLPSEE970 pKa = 4.63EE971 pKa = 4.23RR972 pKa = 11.84YY973 pKa = 10.47SYY975 pKa = 11.02LFEE978 pKa = 5.53GNAQAFDD985 pKa = 4.42NILVSGGLRR994 pKa = 11.84NNAQFDD1000 pKa = 4.1VVHH1003 pKa = 6.21YY1004 pKa = 10.73NSEE1007 pKa = 3.86QLAANQATDD1016 pKa = 3.14HH1017 pKa = 6.59DD1018 pKa = 4.4QVVARR1023 pKa = 11.84IEE1025 pKa = 3.96IPLPNVAPSGADD1037 pKa = 3.02ASRR1040 pKa = 11.84TILEE1044 pKa = 4.36DD1045 pKa = 3.1ASYY1048 pKa = 10.11TFATADD1054 pKa = 3.51FGFADD1059 pKa = 4.1GNGNALSGVRR1069 pKa = 11.84ITTLPGAGTLFLDD1082 pKa = 4.45ADD1084 pKa = 4.34GAGGNAPVVVTAGQTISAADD1104 pKa = 3.3IAAGKK1109 pKa = 8.56LTFVPAADD1117 pKa = 4.18GNGAGYY1123 pKa = 10.94GSFTFQVQDD1132 pKa = 4.29DD1133 pKa = 4.35GGTVNGGVNLDD1144 pKa = 3.41QSANSFTVNVTAQDD1158 pKa = 3.85DD1159 pKa = 4.24APTAVADD1166 pKa = 3.86VVTTAEE1172 pKa = 4.1NIIAVVNVVGNDD1184 pKa = 3.15TDD1186 pKa = 3.76IDD1188 pKa = 4.05GGPKK1192 pKa = 10.3GVVSVNGNALTSGQSTTLASGAIVTLGDD1220 pKa = 4.51DD1221 pKa = 3.4GTLRR1225 pKa = 11.84YY1226 pKa = 10.06DD1227 pKa = 3.49PNGAFNGLSTGTTSADD1243 pKa = 3.13SFTYY1247 pKa = 10.81ALNGGSAASVSVTINGVTQVTGDD1270 pKa = 3.71SGDD1273 pKa = 3.59NNQAGTGSADD1283 pKa = 3.1NYY1285 pKa = 10.83NFSQGGDD1292 pKa = 3.59DD1293 pKa = 3.67TVSTGGGSDD1302 pKa = 4.0AIFFGAAFNQNDD1314 pKa = 4.33KK1315 pKa = 11.13VDD1317 pKa = 3.98GGAGNNDD1324 pKa = 3.25QLGLQGDD1331 pKa = 4.28YY1332 pKa = 10.99TGGNALTLGASNVTGVEE1349 pKa = 4.4VVALLAGFDD1358 pKa = 4.19YY1359 pKa = 11.32NVTTLDD1365 pKa = 3.94DD1366 pKa = 4.84LLTTGQTMTVFGGGLGAGDD1385 pKa = 4.25NLTFDD1390 pKa = 4.63GSAEE1394 pKa = 4.02TSGNFLVYY1402 pKa = 10.49GGLGTDD1408 pKa = 3.44VLTGGGGNDD1417 pKa = 3.46GFSFGAGRR1425 pKa = 11.84FNAATDD1431 pKa = 3.71VVNGGTGADD1440 pKa = 3.19QVALNGFTGTVSGTNFISVEE1460 pKa = 4.33TIVLATGSNTVTLADD1475 pKa = 3.72DD1476 pKa = 3.45WTLGGEE1482 pKa = 4.11SHH1484 pKa = 6.22TVFGSALSQGVTVDD1498 pKa = 4.4GSAEE1502 pKa = 4.0TDD1504 pKa = 3.1GRR1506 pKa = 11.84LALYY1510 pKa = 10.04GGSGNDD1516 pKa = 4.04LLTGGAGNDD1525 pKa = 4.21FIVGGAGADD1534 pKa = 3.86TIRR1537 pKa = 11.84GGLGADD1543 pKa = 4.38LLRR1546 pKa = 11.84GDD1548 pKa = 4.52GGADD1552 pKa = 3.24TFVLGSVAEE1561 pKa = 4.53STSTTHH1567 pKa = 6.63DD1568 pKa = 3.34TLIGFDD1574 pKa = 3.58VSADD1578 pKa = 3.84KK1579 pKa = 10.76IDD1581 pKa = 4.03LSTTITAIDD1590 pKa = 3.39AAITVGTLSNGSFDD1604 pKa = 4.67SDD1606 pKa = 3.74LTTALAGLGANHH1618 pKa = 8.01AITFTADD1625 pKa = 2.8QGDD1628 pKa = 3.59LAGRR1632 pKa = 11.84IFLVADD1638 pKa = 3.46TNGIAGYY1645 pKa = 9.18QAGQDD1650 pKa = 3.28AVFEE1654 pKa = 5.21LINPVQPITDD1664 pKa = 3.77PTPFII1669 pKa = 4.85
SS1 pKa = 4.87YY2 pKa = 9.85TFSVVVNGDD11 pKa = 3.47ALVEE15 pKa = 4.26PNEE18 pKa = 4.29TVLVNLSNVVGAAVTDD34 pKa = 3.98SQGSGTIIDD43 pKa = 3.92NDD45 pKa = 3.88FPTLSISDD53 pKa = 3.8ASVSEE58 pKa = 4.35GDD60 pKa = 3.35SGTTVLTYY68 pKa = 10.13TVTASAPALAGGISFTIATADD89 pKa = 3.48GTATAGSDD97 pKa = 3.29YY98 pKa = 10.83DD99 pKa = 3.86AQSTTGTIAEE109 pKa = 4.4GQTTYY114 pKa = 10.66TFSVVVNGDD123 pKa = 3.13VSNEE127 pKa = 4.0SNEE130 pKa = 4.24TVLVNLSNVTGATLVDD146 pKa = 3.68AQGVGTIVNDD156 pKa = 4.17DD157 pKa = 3.49GPIRR161 pKa = 11.84IFSEE165 pKa = 4.14SFSNFLAAGFAPTPSASQLDD185 pKa = 3.71SDD187 pKa = 3.82IWRR190 pKa = 11.84VAGLSDD196 pKa = 4.13NLTPAYY202 pKa = 10.0GATLTTGDD210 pKa = 3.88FARR213 pKa = 11.84GVINGAADD221 pKa = 3.59PTSGGVYY228 pKa = 10.14SPSANRR234 pKa = 11.84ALVVQPTGSDD244 pKa = 3.19FDD246 pKa = 3.98AGGFIEE252 pKa = 5.95AKK254 pKa = 9.43IQNSSGKK261 pKa = 7.61TAVAFDD267 pKa = 3.92VGFDD271 pKa = 3.0WAYY274 pKa = 10.6RR275 pKa = 11.84NSGNRR280 pKa = 11.84ADD282 pKa = 4.92NMQFSYY288 pKa = 9.61STDD291 pKa = 2.97GTNFTVVPAAAFVTPAAADD310 pKa = 3.47ASVAATFSNQNEE322 pKa = 4.61TVSLTGLAVAAGGFLYY338 pKa = 10.68LRR340 pKa = 11.84WTHH343 pKa = 6.86LDD345 pKa = 3.21GATGSGNRR353 pKa = 11.84DD354 pKa = 3.33EE355 pKa = 5.49IGIDD359 pKa = 3.23NVTIDD364 pKa = 3.64ATTSNLATATVANISLAEE382 pKa = 4.24GDD384 pKa = 4.3AGTTSATFTVVRR396 pKa = 11.84SSGVGAASIDD406 pKa = 3.73YY407 pKa = 7.65TTANGSAIAGTDD419 pKa = 3.46YY420 pKa = 10.96VAASGTVSFADD431 pKa = 3.92GEE433 pKa = 4.53TSQTVTILITGDD445 pKa = 3.33TAFEE449 pKa = 4.18AGEE452 pKa = 4.35TFFLNLSNPSNVLLPSSQASVTITNDD478 pKa = 3.02DD479 pKa = 3.66NGAVAIYY486 pKa = 9.47DD487 pKa = 3.55IQGAGHH493 pKa = 6.58TSALVGQFIQTSGIVTAVAANGFYY517 pKa = 10.83LQDD520 pKa = 3.49PTGDD524 pKa = 3.5GNTRR528 pKa = 11.84TSDD531 pKa = 3.75GIFVFTSTAPTVVVGNGISLTGTVTEE557 pKa = 5.06FDD559 pKa = 3.91AGLDD563 pKa = 3.78TLTVTQITAPTITVLTQTNTLPNAVLIGATGGILPPTQSIDD604 pKa = 3.3SDD606 pKa = 3.66NFTVFNPEE614 pKa = 2.83VDD616 pKa = 4.59GIDD619 pKa = 3.57FYY621 pKa = 11.46EE622 pKa = 4.26TLEE625 pKa = 4.08GMRR628 pKa = 11.84VTVQAPTVTSNTNSFGEE645 pKa = 4.32TFIVASGGIGATGVNEE661 pKa = 4.06RR662 pKa = 11.84GGITISAGDD671 pKa = 3.91NNPEE675 pKa = 4.77RR676 pKa = 11.84IQIDD680 pKa = 3.39NDD682 pKa = 3.63TTIFAGYY689 pKa = 7.75TPSHH693 pKa = 5.72TVGDD697 pKa = 3.81RR698 pKa = 11.84LGDD701 pKa = 3.4VTGVFNYY708 pKa = 10.25AFQSYY713 pKa = 8.46EE714 pKa = 4.28LIVTEE719 pKa = 4.37TVVVTNDD726 pKa = 3.26TTPTRR731 pKa = 11.84EE732 pKa = 4.04TTTLAGDD739 pKa = 3.62ATHH742 pKa = 5.97VTFASFNIEE751 pKa = 4.04NADD754 pKa = 3.6PTDD757 pKa = 3.97PASKK761 pKa = 8.98FTLLGTEE768 pKa = 3.57IRR770 pKa = 11.84YY771 pKa = 9.35ALNTPDD777 pKa = 4.83IIALQEE783 pKa = 3.93VQDD786 pKa = 4.09ADD788 pKa = 3.93GAGTGTNYY796 pKa = 10.61SGTVTAQKK804 pKa = 10.71LIDD807 pKa = 4.78AIIAAGGPTYY817 pKa = 10.61SYY819 pKa = 11.28IEE821 pKa = 4.08IAPTANNTTGGEE833 pKa = 4.06PNGNIRR839 pKa = 11.84NGFLYY844 pKa = 10.2NAARR848 pKa = 11.84VTYY851 pKa = 10.35VDD853 pKa = 3.54GSVALVPGIAYY864 pKa = 10.04NNSRR868 pKa = 11.84NPLVADD874 pKa = 4.74FSFNGQVFTTVSVHH888 pKa = 4.12STSRR892 pKa = 11.84GGSDD896 pKa = 3.47AFYY899 pKa = 11.29GDD901 pKa = 3.42IQPPVQAGDD910 pKa = 3.75SQRR913 pKa = 11.84TAQADD918 pKa = 3.69TLRR921 pKa = 11.84AYY923 pKa = 9.89INDD926 pKa = 3.69QLATDD931 pKa = 4.82PNHH934 pKa = 5.82QFIIGGDD941 pKa = 3.61FNGYY945 pKa = 9.52YY946 pKa = 9.9YY947 pKa = 8.81EE948 pKa = 4.1TAMQRR953 pKa = 11.84LVAGGVLTNLYY964 pKa = 10.5DD965 pKa = 4.21LLPSEE970 pKa = 4.63EE971 pKa = 4.23RR972 pKa = 11.84YY973 pKa = 10.47SYY975 pKa = 11.02LFEE978 pKa = 5.53GNAQAFDD985 pKa = 4.42NILVSGGLRR994 pKa = 11.84NNAQFDD1000 pKa = 4.1VVHH1003 pKa = 6.21YY1004 pKa = 10.73NSEE1007 pKa = 3.86QLAANQATDD1016 pKa = 3.14HH1017 pKa = 6.59DD1018 pKa = 4.4QVVARR1023 pKa = 11.84IEE1025 pKa = 3.96IPLPNVAPSGADD1037 pKa = 3.02ASRR1040 pKa = 11.84TILEE1044 pKa = 4.36DD1045 pKa = 3.1ASYY1048 pKa = 10.11TFATADD1054 pKa = 3.51FGFADD1059 pKa = 4.1GNGNALSGVRR1069 pKa = 11.84ITTLPGAGTLFLDD1082 pKa = 4.45ADD1084 pKa = 4.34GAGGNAPVVVTAGQTISAADD1104 pKa = 3.3IAAGKK1109 pKa = 8.56LTFVPAADD1117 pKa = 4.18GNGAGYY1123 pKa = 10.94GSFTFQVQDD1132 pKa = 4.29DD1133 pKa = 4.35GGTVNGGVNLDD1144 pKa = 3.41QSANSFTVNVTAQDD1158 pKa = 3.85DD1159 pKa = 4.24APTAVADD1166 pKa = 3.86VVTTAEE1172 pKa = 4.1NIIAVVNVVGNDD1184 pKa = 3.15TDD1186 pKa = 3.76IDD1188 pKa = 4.05GGPKK1192 pKa = 10.3GVVSVNGNALTSGQSTTLASGAIVTLGDD1220 pKa = 4.51DD1221 pKa = 3.4GTLRR1225 pKa = 11.84YY1226 pKa = 10.06DD1227 pKa = 3.49PNGAFNGLSTGTTSADD1243 pKa = 3.13SFTYY1247 pKa = 10.81ALNGGSAASVSVTINGVTQVTGDD1270 pKa = 3.71SGDD1273 pKa = 3.59NNQAGTGSADD1283 pKa = 3.1NYY1285 pKa = 10.83NFSQGGDD1292 pKa = 3.59DD1293 pKa = 3.67TVSTGGGSDD1302 pKa = 4.0AIFFGAAFNQNDD1314 pKa = 4.33KK1315 pKa = 11.13VDD1317 pKa = 3.98GGAGNNDD1324 pKa = 3.25QLGLQGDD1331 pKa = 4.28YY1332 pKa = 10.99TGGNALTLGASNVTGVEE1349 pKa = 4.4VVALLAGFDD1358 pKa = 4.19YY1359 pKa = 11.32NVTTLDD1365 pKa = 3.94DD1366 pKa = 4.84LLTTGQTMTVFGGGLGAGDD1385 pKa = 4.25NLTFDD1390 pKa = 4.63GSAEE1394 pKa = 4.02TSGNFLVYY1402 pKa = 10.49GGLGTDD1408 pKa = 3.44VLTGGGGNDD1417 pKa = 3.46GFSFGAGRR1425 pKa = 11.84FNAATDD1431 pKa = 3.71VVNGGTGADD1440 pKa = 3.19QVALNGFTGTVSGTNFISVEE1460 pKa = 4.33TIVLATGSNTVTLADD1475 pKa = 3.72DD1476 pKa = 3.45WTLGGEE1482 pKa = 4.11SHH1484 pKa = 6.22TVFGSALSQGVTVDD1498 pKa = 4.4GSAEE1502 pKa = 4.0TDD1504 pKa = 3.1GRR1506 pKa = 11.84LALYY1510 pKa = 10.04GGSGNDD1516 pKa = 4.04LLTGGAGNDD1525 pKa = 4.21FIVGGAGADD1534 pKa = 3.86TIRR1537 pKa = 11.84GGLGADD1543 pKa = 4.38LLRR1546 pKa = 11.84GDD1548 pKa = 4.52GGADD1552 pKa = 3.24TFVLGSVAEE1561 pKa = 4.53STSTTHH1567 pKa = 6.63DD1568 pKa = 3.34TLIGFDD1574 pKa = 3.58VSADD1578 pKa = 3.84KK1579 pKa = 10.76IDD1581 pKa = 4.03LSTTITAIDD1590 pKa = 3.39AAITVGTLSNGSFDD1604 pKa = 4.67SDD1606 pKa = 3.74LTTALAGLGANHH1618 pKa = 8.01AITFTADD1625 pKa = 2.8QGDD1628 pKa = 3.59LAGRR1632 pKa = 11.84IFLVADD1638 pKa = 3.46TNGIAGYY1645 pKa = 9.18QAGQDD1650 pKa = 3.28AVFEE1654 pKa = 5.21LINPVQPITDD1664 pKa = 3.77PTPFII1669 pKa = 4.85
Molecular weight: 168.87 kDa
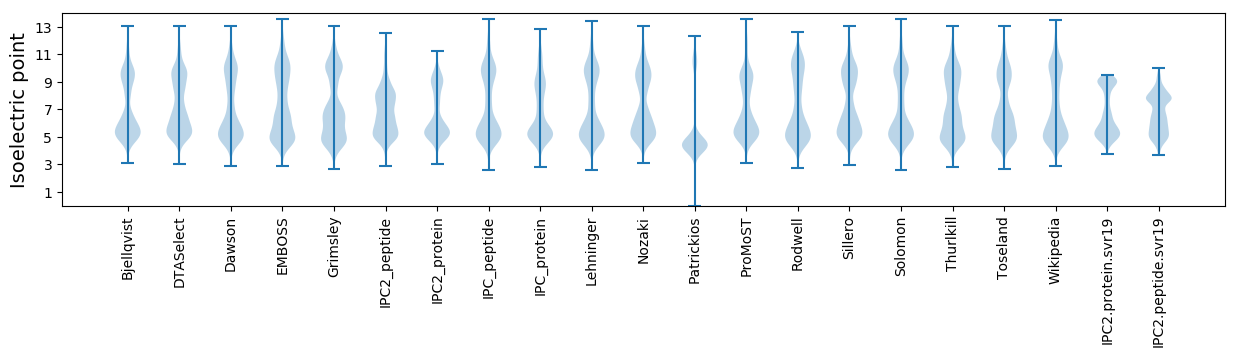
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A2R7IKR1|A0A2R7IKR1_9SPHN RND transporter (Fragment) OS=Sphingomonas sp. HMWF008 OX=2056845 GN=DBR17_18205 PE=3 SV=1
MM1 pKa = 7.45KK2 pKa = 9.61RR3 pKa = 11.84TFQPSNLVRR12 pKa = 11.84ARR14 pKa = 11.84RR15 pKa = 11.84HH16 pKa = 4.58GFRR19 pKa = 11.84SRR21 pKa = 11.84MATPGGRR28 pKa = 11.84NVIRR32 pKa = 11.84ARR34 pKa = 11.84RR35 pKa = 11.84SRR37 pKa = 11.84GRR39 pKa = 11.84KK40 pKa = 8.97KK41 pKa = 10.66LSAA44 pKa = 4.03
MM1 pKa = 7.45KK2 pKa = 9.61RR3 pKa = 11.84TFQPSNLVRR12 pKa = 11.84ARR14 pKa = 11.84RR15 pKa = 11.84HH16 pKa = 4.58GFRR19 pKa = 11.84SRR21 pKa = 11.84MATPGGRR28 pKa = 11.84NVIRR32 pKa = 11.84ARR34 pKa = 11.84RR35 pKa = 11.84SRR37 pKa = 11.84GRR39 pKa = 11.84KK40 pKa = 8.97KK41 pKa = 10.66LSAA44 pKa = 4.03
Molecular weight: 5.12 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
974285 |
12 |
3638 |
275.5 |
29.63 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
13.677 ± 0.063 | 0.73 ± 0.012 |
5.942 ± 0.032 | 4.84 ± 0.041 |
3.618 ± 0.026 | 8.857 ± 0.044 |
1.903 ± 0.023 | 5.132 ± 0.025 |
2.956 ± 0.036 | 9.825 ± 0.045 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.272 ± 0.023 | 2.566 ± 0.033 |
5.36 ± 0.035 | 3.035 ± 0.023 |
7.096 ± 0.041 | 5.241 ± 0.033 |
5.826 ± 0.039 | 7.428 ± 0.037 |
1.41 ± 0.018 | 2.276 ± 0.028 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
