
Pseudomonas pelagia
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Gammaproteobacteria; Pseudomonadales; Pseudomonadaceae; Pseudomonas
Average proteome isoelectric point is 6.13
Get precalculated fractions of proteins
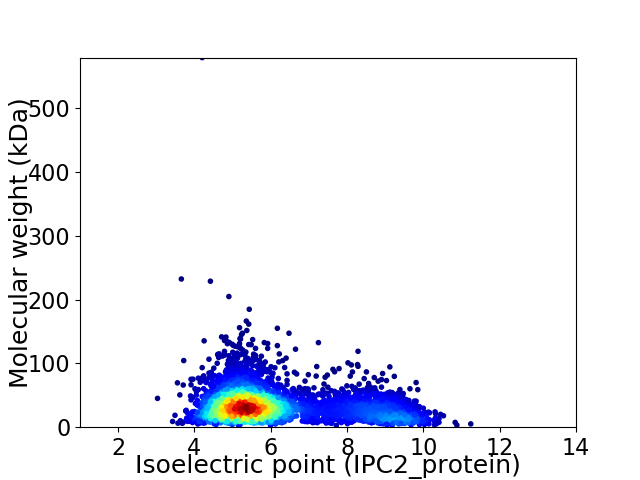
Virtual 2D-PAGE plot for 4145 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A2A4AWA0|A0A2A4AWA0_9PSED LysR family transcriptional regulator OS=Pseudomonas pelagia OX=553151 GN=CO192_07230 PE=3 SV=1
MM1 pKa = 7.29SACLDD6 pKa = 3.62SDD8 pKa = 4.09GGSSSADD15 pKa = 2.98NDD17 pKa = 3.71AAPPPEE23 pKa = 4.07NTKK26 pKa = 10.65PNLILIIADD35 pKa = 3.59DD36 pKa = 3.98VGVDD40 pKa = 3.42QLASFGFGGAEE51 pKa = 3.97PPATPTIDD59 pKa = 3.27SLAATGVSFNNTWSMPTCSTTRR81 pKa = 11.84AALLSGRR88 pKa = 11.84YY89 pKa = 8.63PPRR92 pKa = 11.84TNVNTAIVSTDD103 pKa = 3.62FANSQTSPFEE113 pKa = 3.91RR114 pKa = 11.84SLPTVLGEE122 pKa = 3.91QGYY125 pKa = 9.71VSAYY129 pKa = 8.53VGKK132 pKa = 9.96IHH134 pKa = 6.21VTGSDD139 pKa = 3.62VNPANHH145 pKa = 7.18PFGDD149 pKa = 3.7EE150 pKa = 4.32AIRR153 pKa = 11.84ALGFDD158 pKa = 3.62YY159 pKa = 10.68FAGYY163 pKa = 10.18FDD165 pKa = 4.6GGPRR169 pKa = 11.84PIDD172 pKa = 3.34VTAGLDD178 pKa = 3.68NLDD181 pKa = 4.14SSAAPYY187 pKa = 9.41TCGFVPLATDD197 pKa = 4.1HH198 pKa = 6.64PQGADD203 pKa = 2.86TGACYY208 pKa = 9.92QPSGACEE215 pKa = 4.04EE216 pKa = 4.29LTADD220 pKa = 4.06GSSVPGKK227 pKa = 9.36MCLEE231 pKa = 4.23RR232 pKa = 11.84GGLFDD237 pKa = 6.32PNQQCASTLPAYY249 pKa = 10.9LDD251 pKa = 3.65FTRR254 pKa = 11.84QNAYY258 pKa = 8.27YY259 pKa = 8.35TAEE262 pKa = 4.08VVISEE267 pKa = 4.38EE268 pKa = 4.16SGSQQLEE275 pKa = 3.84VDD277 pKa = 3.6NPATRR282 pKa = 11.84LYY284 pKa = 9.47RR285 pKa = 11.84TQLEE289 pKa = 4.72SNFAIDD295 pKa = 4.31WIQAQSDD302 pKa = 3.91EE303 pKa = 4.46QPWMLSLGYY312 pKa = 9.46SASHH316 pKa = 6.93APLQPTPASLTPEE329 pKa = 4.06TGALDD334 pKa = 4.24IGIACDD340 pKa = 3.31NGADD344 pKa = 3.67TRR346 pKa = 11.84KK347 pKa = 10.3LMNQNMEE354 pKa = 4.21AMDD357 pKa = 3.73TEE359 pKa = 4.06IRR361 pKa = 11.84RR362 pKa = 11.84VLLAAGVYY370 pKa = 9.68IEE372 pKa = 4.64NANGDD377 pKa = 4.18LEE379 pKa = 4.52YY380 pKa = 11.18NPASNTVVAFIGDD393 pKa = 3.61NGTYY397 pKa = 10.45GPVVKK402 pKa = 10.69DD403 pKa = 3.3PFLPARR409 pKa = 11.84SKK411 pKa = 9.79GTVYY415 pKa = 10.44QGGIWVPLIVTGATVEE431 pKa = 4.05QPGRR435 pKa = 11.84SVGAMVNSTDD445 pKa = 4.48LYY447 pKa = 11.22HH448 pKa = 7.49LFATLGGADD457 pKa = 3.85LSAPEE462 pKa = 3.97VADD465 pKa = 4.57RR466 pKa = 11.84LDD468 pKa = 4.15AQPMDD473 pKa = 4.38AYY475 pKa = 10.53LASAEE480 pKa = 4.66AIPIRR485 pKa = 11.84DD486 pKa = 3.79SNFSMSGRR494 pKa = 11.84NLASAIPQPCVLTDD508 pKa = 3.58VNICLQLFPQKK519 pKa = 10.36GLCISEE525 pKa = 3.96GGTWYY530 pKa = 10.78GDD532 pKa = 3.54DD533 pKa = 3.83GVVPGQSFTSCCAVNTYY550 pKa = 9.88RR551 pKa = 11.84VSEE554 pKa = 4.4GEE556 pKa = 4.02APFDD560 pKa = 3.93ILAEE564 pKa = 4.22TQRR567 pKa = 11.84TVRR570 pKa = 11.84DD571 pKa = 3.6DD572 pKa = 3.61RR573 pKa = 11.84FKK575 pKa = 11.22LLQLEE580 pKa = 4.71EE581 pKa = 4.79PNCDD585 pKa = 3.0AGGLTEE591 pKa = 4.79SFEE594 pKa = 4.28FYY596 pKa = 10.5EE597 pKa = 4.38INEE600 pKa = 4.42SGSEE604 pKa = 3.93LGLDD608 pKa = 3.57NLDD611 pKa = 4.04AQDD614 pKa = 4.19LLTRR618 pKa = 11.84SALTAEE624 pKa = 4.46QQQHH628 pKa = 5.78YY629 pKa = 7.97NTLLAEE635 pKa = 4.68LDD637 pKa = 4.19SIVDD641 pKa = 4.04SEE643 pKa = 4.58PTCPGDD649 pKa = 3.58GNLDD653 pKa = 3.53RR654 pKa = 11.84VINEE658 pKa = 3.83QDD660 pKa = 3.48LKK662 pKa = 10.61DD663 pKa = 3.34WSYY666 pKa = 11.45FSQLNGGRR674 pKa = 11.84SSWYY678 pKa = 10.17DD679 pKa = 3.41FNLDD683 pKa = 3.65GQTDD687 pKa = 3.4AADD690 pKa = 3.83RR691 pKa = 11.84QIIEE695 pKa = 4.73DD696 pKa = 3.97NLGLEE701 pKa = 4.68CLL703 pKa = 4.09
MM1 pKa = 7.29SACLDD6 pKa = 3.62SDD8 pKa = 4.09GGSSSADD15 pKa = 2.98NDD17 pKa = 3.71AAPPPEE23 pKa = 4.07NTKK26 pKa = 10.65PNLILIIADD35 pKa = 3.59DD36 pKa = 3.98VGVDD40 pKa = 3.42QLASFGFGGAEE51 pKa = 3.97PPATPTIDD59 pKa = 3.27SLAATGVSFNNTWSMPTCSTTRR81 pKa = 11.84AALLSGRR88 pKa = 11.84YY89 pKa = 8.63PPRR92 pKa = 11.84TNVNTAIVSTDD103 pKa = 3.62FANSQTSPFEE113 pKa = 3.91RR114 pKa = 11.84SLPTVLGEE122 pKa = 3.91QGYY125 pKa = 9.71VSAYY129 pKa = 8.53VGKK132 pKa = 9.96IHH134 pKa = 6.21VTGSDD139 pKa = 3.62VNPANHH145 pKa = 7.18PFGDD149 pKa = 3.7EE150 pKa = 4.32AIRR153 pKa = 11.84ALGFDD158 pKa = 3.62YY159 pKa = 10.68FAGYY163 pKa = 10.18FDD165 pKa = 4.6GGPRR169 pKa = 11.84PIDD172 pKa = 3.34VTAGLDD178 pKa = 3.68NLDD181 pKa = 4.14SSAAPYY187 pKa = 9.41TCGFVPLATDD197 pKa = 4.1HH198 pKa = 6.64PQGADD203 pKa = 2.86TGACYY208 pKa = 9.92QPSGACEE215 pKa = 4.04EE216 pKa = 4.29LTADD220 pKa = 4.06GSSVPGKK227 pKa = 9.36MCLEE231 pKa = 4.23RR232 pKa = 11.84GGLFDD237 pKa = 6.32PNQQCASTLPAYY249 pKa = 10.9LDD251 pKa = 3.65FTRR254 pKa = 11.84QNAYY258 pKa = 8.27YY259 pKa = 8.35TAEE262 pKa = 4.08VVISEE267 pKa = 4.38EE268 pKa = 4.16SGSQQLEE275 pKa = 3.84VDD277 pKa = 3.6NPATRR282 pKa = 11.84LYY284 pKa = 9.47RR285 pKa = 11.84TQLEE289 pKa = 4.72SNFAIDD295 pKa = 4.31WIQAQSDD302 pKa = 3.91EE303 pKa = 4.46QPWMLSLGYY312 pKa = 9.46SASHH316 pKa = 6.93APLQPTPASLTPEE329 pKa = 4.06TGALDD334 pKa = 4.24IGIACDD340 pKa = 3.31NGADD344 pKa = 3.67TRR346 pKa = 11.84KK347 pKa = 10.3LMNQNMEE354 pKa = 4.21AMDD357 pKa = 3.73TEE359 pKa = 4.06IRR361 pKa = 11.84RR362 pKa = 11.84VLLAAGVYY370 pKa = 9.68IEE372 pKa = 4.64NANGDD377 pKa = 4.18LEE379 pKa = 4.52YY380 pKa = 11.18NPASNTVVAFIGDD393 pKa = 3.61NGTYY397 pKa = 10.45GPVVKK402 pKa = 10.69DD403 pKa = 3.3PFLPARR409 pKa = 11.84SKK411 pKa = 9.79GTVYY415 pKa = 10.44QGGIWVPLIVTGATVEE431 pKa = 4.05QPGRR435 pKa = 11.84SVGAMVNSTDD445 pKa = 4.48LYY447 pKa = 11.22HH448 pKa = 7.49LFATLGGADD457 pKa = 3.85LSAPEE462 pKa = 3.97VADD465 pKa = 4.57RR466 pKa = 11.84LDD468 pKa = 4.15AQPMDD473 pKa = 4.38AYY475 pKa = 10.53LASAEE480 pKa = 4.66AIPIRR485 pKa = 11.84DD486 pKa = 3.79SNFSMSGRR494 pKa = 11.84NLASAIPQPCVLTDD508 pKa = 3.58VNICLQLFPQKK519 pKa = 10.36GLCISEE525 pKa = 3.96GGTWYY530 pKa = 10.78GDD532 pKa = 3.54DD533 pKa = 3.83GVVPGQSFTSCCAVNTYY550 pKa = 9.88RR551 pKa = 11.84VSEE554 pKa = 4.4GEE556 pKa = 4.02APFDD560 pKa = 3.93ILAEE564 pKa = 4.22TQRR567 pKa = 11.84TVRR570 pKa = 11.84DD571 pKa = 3.6DD572 pKa = 3.61RR573 pKa = 11.84FKK575 pKa = 11.22LLQLEE580 pKa = 4.71EE581 pKa = 4.79PNCDD585 pKa = 3.0AGGLTEE591 pKa = 4.79SFEE594 pKa = 4.28FYY596 pKa = 10.5EE597 pKa = 4.38INEE600 pKa = 4.42SGSEE604 pKa = 3.93LGLDD608 pKa = 3.57NLDD611 pKa = 4.04AQDD614 pKa = 4.19LLTRR618 pKa = 11.84SALTAEE624 pKa = 4.46QQQHH628 pKa = 5.78YY629 pKa = 7.97NTLLAEE635 pKa = 4.68LDD637 pKa = 4.19SIVDD641 pKa = 4.04SEE643 pKa = 4.58PTCPGDD649 pKa = 3.58GNLDD653 pKa = 3.53RR654 pKa = 11.84VINEE658 pKa = 3.83QDD660 pKa = 3.48LKK662 pKa = 10.61DD663 pKa = 3.34WSYY666 pKa = 11.45FSQLNGGRR674 pKa = 11.84SSWYY678 pKa = 10.17DD679 pKa = 3.41FNLDD683 pKa = 3.65GQTDD687 pKa = 3.4AADD690 pKa = 3.83RR691 pKa = 11.84QIIEE695 pKa = 4.73DD696 pKa = 3.97NLGLEE701 pKa = 4.68CLL703 pKa = 4.09
Molecular weight: 75.08 kDa
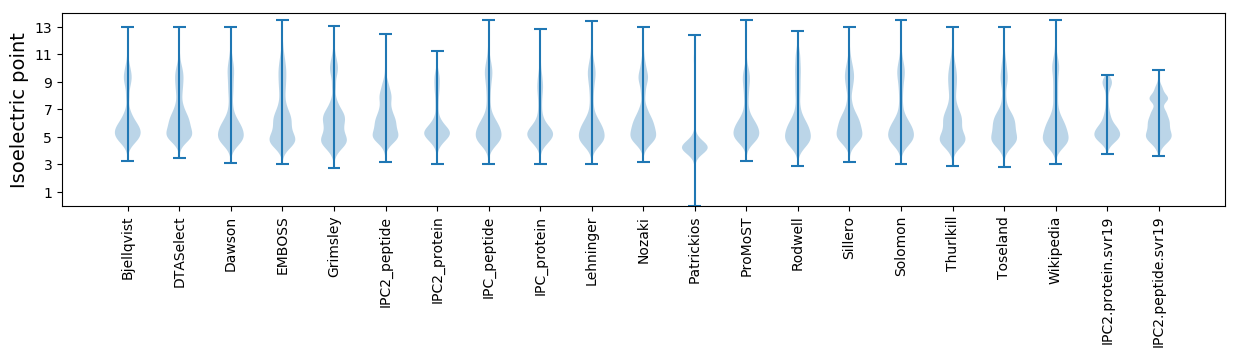
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A2A4AYQ5|A0A2A4AYQ5_9PSED YkuD domain-containing protein OS=Pseudomonas pelagia OX=553151 GN=CO192_00945 PE=3 SV=1
MM1 pKa = 7.45KK2 pKa = 9.51RR3 pKa = 11.84TFQPSVLKK11 pKa = 10.46RR12 pKa = 11.84KK13 pKa = 9.37RR14 pKa = 11.84NHH16 pKa = 5.37GFRR19 pKa = 11.84ARR21 pKa = 11.84MATANGRR28 pKa = 11.84NVLARR33 pKa = 11.84RR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 9.93GRR39 pKa = 11.84KK40 pKa = 8.85RR41 pKa = 11.84LSAA44 pKa = 3.96
MM1 pKa = 7.45KK2 pKa = 9.51RR3 pKa = 11.84TFQPSVLKK11 pKa = 10.46RR12 pKa = 11.84KK13 pKa = 9.37RR14 pKa = 11.84NHH16 pKa = 5.37GFRR19 pKa = 11.84ARR21 pKa = 11.84MATANGRR28 pKa = 11.84NVLARR33 pKa = 11.84RR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 9.93GRR39 pKa = 11.84KK40 pKa = 8.85RR41 pKa = 11.84LSAA44 pKa = 3.96
Molecular weight: 5.15 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1348114 |
23 |
5694 |
325.2 |
35.77 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
10.527 ± 0.047 | 1.015 ± 0.014 |
5.567 ± 0.032 | 6.021 ± 0.036 |
3.607 ± 0.021 | 7.778 ± 0.043 |
2.258 ± 0.023 | 5.143 ± 0.03 |
3.346 ± 0.033 | 11.48 ± 0.061 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.639 ± 0.02 | 3.11 ± 0.029 |
4.718 ± 0.028 | 4.649 ± 0.031 |
6.297 ± 0.038 | 5.96 ± 0.033 |
4.901 ± 0.026 | 7.033 ± 0.035 |
1.426 ± 0.016 | 2.525 ± 0.019 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
