
Gammapapillomavirus 22
Taxonomy: Viruses; Monodnaviria; Shotokuvirae; Cossaviricota; Papovaviricetes; Zurhausenvirales; Papillomaviridae; Firstpapillomavirinae; Gammapapillomavirus
Average proteome isoelectric point is 6.4
Get precalculated fractions of proteins

Virtual 2D-PAGE plot for 7 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
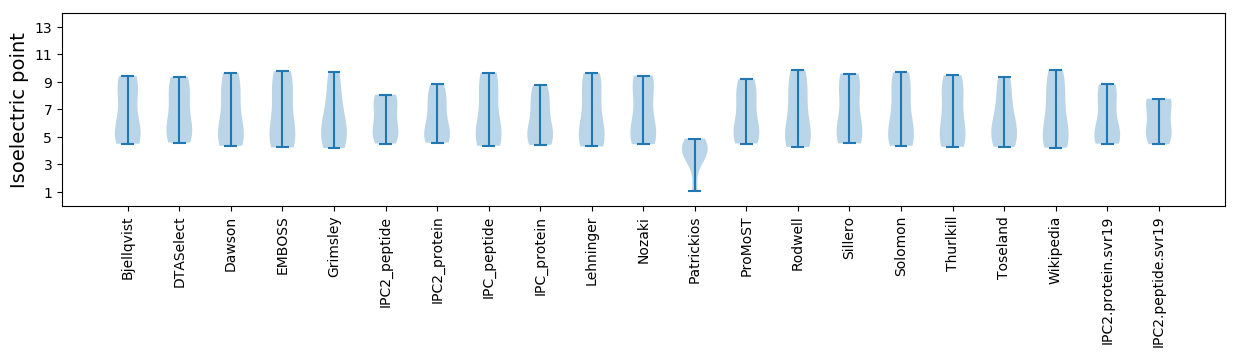
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions

Protein with the lowest isoelectric point:
>tr|A0A2D2AM26|A0A2D2AM26_9PAPI E4 OS=Gammapapillomavirus 22 OX=1961679 GN=E4 PE=4 SV=1
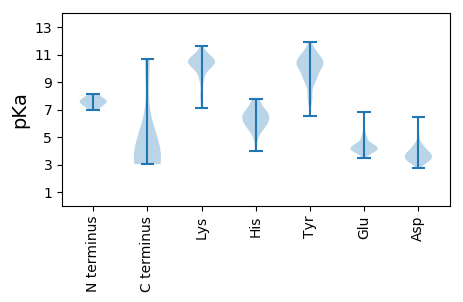
MM1 pKa = 7.75IGPDD5 pKa = 3.46VKK7 pKa = 11.1SGDD10 pKa = 3.4IEE12 pKa = 5.61LDD14 pKa = 3.55LEE16 pKa = 4.64SLVLPEE22 pKa = 4.58NLLSNEE28 pKa = 4.07SLSPDD33 pKa = 3.38TEE35 pKa = 4.33GQAEE39 pKa = 4.3EE40 pKa = 5.04VEE42 pKa = 4.29QAPYY46 pKa = 9.96TVDD49 pKa = 3.65TSCNSCGARR58 pKa = 11.84VRR60 pKa = 11.84ICVIATRR67 pKa = 11.84IAILTLQEE75 pKa = 4.54LLTLEE80 pKa = 5.33LNLLCPPCSRR90 pKa = 11.84TVVRR94 pKa = 11.84HH95 pKa = 4.57GRR97 pKa = 11.84RR98 pKa = 3.23
MM1 pKa = 7.75IGPDD5 pKa = 3.46VKK7 pKa = 11.1SGDD10 pKa = 3.4IEE12 pKa = 5.61LDD14 pKa = 3.55LEE16 pKa = 4.64SLVLPEE22 pKa = 4.58NLLSNEE28 pKa = 4.07SLSPDD33 pKa = 3.38TEE35 pKa = 4.33GQAEE39 pKa = 4.3EE40 pKa = 5.04VEE42 pKa = 4.29QAPYY46 pKa = 9.96TVDD49 pKa = 3.65TSCNSCGARR58 pKa = 11.84VRR60 pKa = 11.84ICVIATRR67 pKa = 11.84IAILTLQEE75 pKa = 4.54LLTLEE80 pKa = 5.33LNLLCPPCSRR90 pKa = 11.84TVVRR94 pKa = 11.84HH95 pKa = 4.57GRR97 pKa = 11.84RR98 pKa = 3.23
Molecular weight: 10.69 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A2D2AM35|A0A2D2AM35_9PAPI Replication protein E1 OS=Gammapapillomavirus 22 OX=1961679 GN=E1 PE=3 SV=1
MM1 pKa = 7.0NQADD5 pKa = 3.78LTDD8 pKa = 4.84RR9 pKa = 11.84YY10 pKa = 10.82DD11 pKa = 3.71ALQEE15 pKa = 4.08RR16 pKa = 11.84LMSLYY21 pKa = 10.3EE22 pKa = 3.95EE23 pKa = 4.38SSNTIEE29 pKa = 4.01AQIMLWEE36 pKa = 4.93TIRR39 pKa = 11.84KK40 pKa = 7.83EE41 pKa = 4.22QVLLYY46 pKa = 9.47YY47 pKa = 10.39GRR49 pKa = 11.84KK50 pKa = 9.15EE51 pKa = 4.0GYY53 pKa = 10.09KK54 pKa = 10.54NFGLQPIPNLAVSEE68 pKa = 4.21YY69 pKa = 9.39RR70 pKa = 11.84AKK72 pKa = 10.67EE73 pKa = 4.1AIQQVLLLKK82 pKa = 10.41SLQKK86 pKa = 10.5SAFGRR91 pKa = 11.84EE92 pKa = 3.9EE93 pKa = 3.64WTLTDD98 pKa = 3.54TSAEE102 pKa = 4.18LTHH105 pKa = 6.14TAPRR109 pKa = 11.84NTFKK113 pKa = 10.96KK114 pKa = 10.6GPFTVNVYY122 pKa = 10.44FDD124 pKa = 4.54HH125 pKa = 6.76NANNVFPYY133 pKa = 8.29TQWNWLYY140 pKa = 11.11VQDD143 pKa = 5.76DD144 pKa = 3.32NDD146 pKa = 3.35MWYY149 pKa = 7.88KK150 pKa = 10.13TPGLVDD156 pKa = 3.13INGLYY161 pKa = 10.6FEE163 pKa = 5.85DD164 pKa = 4.3NSGEE168 pKa = 4.08KK169 pKa = 10.57NYY171 pKa = 10.46FLLFATDD178 pKa = 3.25AEE180 pKa = 5.03TYY182 pKa = 7.43GTSGGWTVKK191 pKa = 10.52YY192 pKa = 10.51KK193 pKa = 11.08NEE195 pKa = 4.44TISTSTPVTSSQQRR209 pKa = 11.84SFSDD213 pKa = 3.39SFEE216 pKa = 4.36GSSKK220 pKa = 10.96GSVSSSGDD228 pKa = 3.33AVPLPKK234 pKa = 9.47TARR237 pKa = 11.84RR238 pKa = 11.84KK239 pKa = 7.23EE240 pKa = 4.32TEE242 pKa = 3.56EE243 pKa = 4.06GRR245 pKa = 11.84PHH247 pKa = 6.02STTPTTFTVRR257 pKa = 11.84RR258 pKa = 11.84RR259 pKa = 11.84RR260 pKa = 11.84RR261 pKa = 11.84RR262 pKa = 11.84IQQRR266 pKa = 11.84EE267 pKa = 3.87RR268 pKa = 11.84GTTTVSLRR276 pKa = 11.84SEE278 pKa = 3.8RR279 pKa = 11.84RR280 pKa = 11.84RR281 pKa = 11.84TQEE284 pKa = 3.52SQLGVPAGEE293 pKa = 4.48VGRR296 pKa = 11.84GHH298 pKa = 6.76HH299 pKa = 5.95LVPRR303 pKa = 11.84TGLKK307 pKa = 10.08RR308 pKa = 11.84LEE310 pKa = 4.1RR311 pKa = 11.84LEE313 pKa = 5.52AEE315 pKa = 4.5AGDD318 pKa = 3.93PPVILVKK325 pKa = 10.83GSANSLKK332 pKa = 9.97CWRR335 pKa = 11.84NRR337 pKa = 11.84SYY339 pKa = 10.94KK340 pKa = 11.01ANVPCVKK347 pKa = 9.8MSTVFRR353 pKa = 11.84WAEE356 pKa = 3.85CTEE359 pKa = 3.98NNIGANNRR367 pKa = 11.84MLIAFKK373 pKa = 10.79NRR375 pKa = 11.84AQRR378 pKa = 11.84EE379 pKa = 4.39VFLKK383 pKa = 9.47TVRR386 pKa = 11.84FPKK389 pKa = 10.34GVSYY393 pKa = 11.02CFGHH397 pKa = 7.59LDD399 pKa = 3.63SLL401 pKa = 4.38
MM1 pKa = 7.0NQADD5 pKa = 3.78LTDD8 pKa = 4.84RR9 pKa = 11.84YY10 pKa = 10.82DD11 pKa = 3.71ALQEE15 pKa = 4.08RR16 pKa = 11.84LMSLYY21 pKa = 10.3EE22 pKa = 3.95EE23 pKa = 4.38SSNTIEE29 pKa = 4.01AQIMLWEE36 pKa = 4.93TIRR39 pKa = 11.84KK40 pKa = 7.83EE41 pKa = 4.22QVLLYY46 pKa = 9.47YY47 pKa = 10.39GRR49 pKa = 11.84KK50 pKa = 9.15EE51 pKa = 4.0GYY53 pKa = 10.09KK54 pKa = 10.54NFGLQPIPNLAVSEE68 pKa = 4.21YY69 pKa = 9.39RR70 pKa = 11.84AKK72 pKa = 10.67EE73 pKa = 4.1AIQQVLLLKK82 pKa = 10.41SLQKK86 pKa = 10.5SAFGRR91 pKa = 11.84EE92 pKa = 3.9EE93 pKa = 3.64WTLTDD98 pKa = 3.54TSAEE102 pKa = 4.18LTHH105 pKa = 6.14TAPRR109 pKa = 11.84NTFKK113 pKa = 10.96KK114 pKa = 10.6GPFTVNVYY122 pKa = 10.44FDD124 pKa = 4.54HH125 pKa = 6.76NANNVFPYY133 pKa = 8.29TQWNWLYY140 pKa = 11.11VQDD143 pKa = 5.76DD144 pKa = 3.32NDD146 pKa = 3.35MWYY149 pKa = 7.88KK150 pKa = 10.13TPGLVDD156 pKa = 3.13INGLYY161 pKa = 10.6FEE163 pKa = 5.85DD164 pKa = 4.3NSGEE168 pKa = 4.08KK169 pKa = 10.57NYY171 pKa = 10.46FLLFATDD178 pKa = 3.25AEE180 pKa = 5.03TYY182 pKa = 7.43GTSGGWTVKK191 pKa = 10.52YY192 pKa = 10.51KK193 pKa = 11.08NEE195 pKa = 4.44TISTSTPVTSSQQRR209 pKa = 11.84SFSDD213 pKa = 3.39SFEE216 pKa = 4.36GSSKK220 pKa = 10.96GSVSSSGDD228 pKa = 3.33AVPLPKK234 pKa = 9.47TARR237 pKa = 11.84RR238 pKa = 11.84KK239 pKa = 7.23EE240 pKa = 4.32TEE242 pKa = 3.56EE243 pKa = 4.06GRR245 pKa = 11.84PHH247 pKa = 6.02STTPTTFTVRR257 pKa = 11.84RR258 pKa = 11.84RR259 pKa = 11.84RR260 pKa = 11.84RR261 pKa = 11.84RR262 pKa = 11.84IQQRR266 pKa = 11.84EE267 pKa = 3.87RR268 pKa = 11.84GTTTVSLRR276 pKa = 11.84SEE278 pKa = 3.8RR279 pKa = 11.84RR280 pKa = 11.84RR281 pKa = 11.84TQEE284 pKa = 3.52SQLGVPAGEE293 pKa = 4.48VGRR296 pKa = 11.84GHH298 pKa = 6.76HH299 pKa = 5.95LVPRR303 pKa = 11.84TGLKK307 pKa = 10.08RR308 pKa = 11.84LEE310 pKa = 4.1RR311 pKa = 11.84LEE313 pKa = 5.52AEE315 pKa = 4.5AGDD318 pKa = 3.93PPVILVKK325 pKa = 10.83GSANSLKK332 pKa = 9.97CWRR335 pKa = 11.84NRR337 pKa = 11.84SYY339 pKa = 10.94KK340 pKa = 11.01ANVPCVKK347 pKa = 9.8MSTVFRR353 pKa = 11.84WAEE356 pKa = 3.85CTEE359 pKa = 3.98NNIGANNRR367 pKa = 11.84MLIAFKK373 pKa = 10.79NRR375 pKa = 11.84AQRR378 pKa = 11.84EE379 pKa = 4.39VFLKK383 pKa = 9.47TVRR386 pKa = 11.84FPKK389 pKa = 10.34GVSYY393 pKa = 11.02CFGHH397 pKa = 7.59LDD399 pKa = 3.63SLL401 pKa = 4.38
Molecular weight: 45.86 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
2436 |
98 |
601 |
348.0 |
39.51 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
5.337 ± 0.282 | 2.545 ± 0.824 |
6.117 ± 0.527 | 6.568 ± 0.374 |
4.475 ± 0.514 | 4.885 ± 0.505 |
1.642 ± 0.161 | 5.09 ± 0.604 |
5.952 ± 0.989 | 9.278 ± 1.095 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.519 ± 0.347 | 5.172 ± 0.328 |
6.486 ± 0.978 | 3.941 ± 0.388 |
5.624 ± 0.714 | 6.856 ± 0.921 |
7.266 ± 1.168 | 6.199 ± 0.957 |
1.273 ± 0.264 | 3.777 ± 0.656 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
