
Porcine associated porprismacovirus 9
Taxonomy: Viruses; Monodnaviria; Shotokuvirae; Cressdnaviricota; Arfiviricetes; Cremevirales; Smacoviridae; Porprismacovirus
Average proteome isoelectric point is 6.81
Get precalculated fractions of proteins
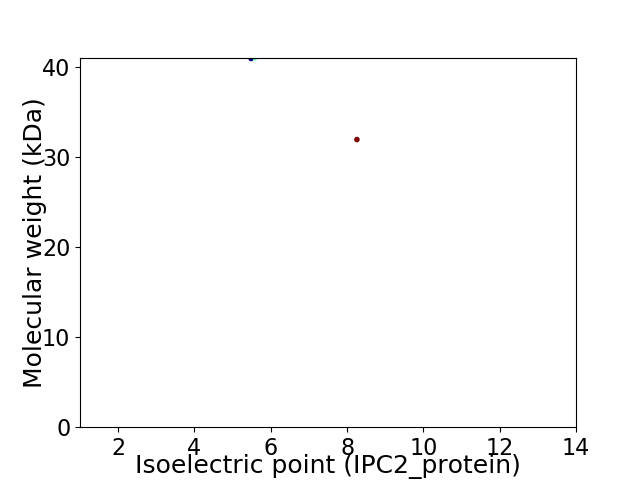
Virtual 2D-PAGE plot for 2 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
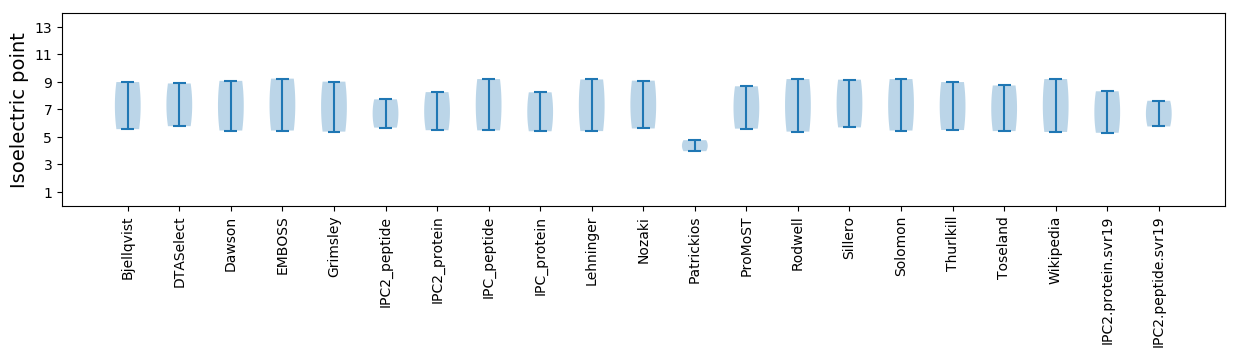
Protein with the lowest isoelectric point:
>tr|A0A076VEP0|A0A076VEP0_9VIRU Cap protein OS=Porcine associated porprismacovirus 9 OX=2170125 PE=4 SV=1
MM1 pKa = 7.26ICIPRR6 pKa = 11.84KK7 pKa = 10.16AFLLCWSFLLLMVFVLLGILLGFFAQFRR35 pKa = 11.84KK36 pKa = 9.6FRR38 pKa = 11.84YY39 pKa = 9.49AGAKK43 pKa = 7.69VTFIPAATLPADD55 pKa = 4.06PLQVSYY61 pKa = 10.83EE62 pKa = 4.08AGEE65 pKa = 4.0PTIDD69 pKa = 5.06PRR71 pKa = 11.84DD72 pKa = 3.66LLNPILHH79 pKa = 6.5KK80 pKa = 10.66GIHH83 pKa = 6.41GDD85 pKa = 3.29ALGNLLEE92 pKa = 4.73EE93 pKa = 4.58YY94 pKa = 10.34KK95 pKa = 10.56QVLASGSSLEE105 pKa = 4.16LTKK108 pKa = 10.2TAAANYY114 pKa = 5.74MTLYY118 pKa = 8.82YY119 pKa = 10.33QCLTDD124 pKa = 4.1PSFKK128 pKa = 10.54KK129 pKa = 10.47SGLRR133 pKa = 11.84QGFVQNLYY141 pKa = 9.99PLVYY145 pKa = 10.58SLGTNRR151 pKa = 11.84QIMPSRR157 pKa = 11.84IFTAGASGLTVGDD170 pKa = 3.99PSLMEE175 pKa = 3.84TGKK178 pKa = 10.16IFSEE182 pKa = 4.09TDD184 pKa = 2.99GQFEE188 pKa = 4.8YY189 pKa = 11.17NDD191 pKa = 3.78EE192 pKa = 4.21TWGVSVRR199 pKa = 11.84RR200 pKa = 11.84SLGQAQQNLKK210 pKa = 10.96DD211 pKa = 4.51NAIEE215 pKa = 3.89WKK217 pKa = 9.37TYY219 pKa = 11.08DD220 pKa = 3.76EE221 pKa = 4.68FTTKK225 pKa = 9.01PTRR228 pKa = 11.84LGWLDD233 pKa = 3.31TLQIIRR239 pKa = 11.84DD240 pKa = 4.14NQSSMPDD247 pKa = 3.32SQVTGVNIEE256 pKa = 3.82LSIRR260 pKa = 11.84NPQDD264 pKa = 3.21PLASRR269 pKa = 11.84VTPCNFTTLPKK280 pKa = 10.03IPMYY284 pKa = 10.3MCFLPPAYY292 pKa = 8.44KK293 pKa = 9.61TEE295 pKa = 3.81MYY297 pKa = 10.14FRR299 pKa = 11.84IVIQHH304 pKa = 5.02YY305 pKa = 10.47FEE307 pKa = 4.84FKK309 pKa = 10.34EE310 pKa = 4.12FRR312 pKa = 11.84STFNQFGIGQVSTATDD328 pKa = 3.55VPYY331 pKa = 11.05DD332 pKa = 3.4WLNAIKK338 pKa = 10.12KK339 pKa = 9.68VSTASVEE346 pKa = 4.4SEE348 pKa = 4.08KK349 pKa = 11.27SLDD352 pKa = 3.46IAGGDD357 pKa = 4.0AEE359 pKa = 4.49LTTDD363 pKa = 3.97GVLL366 pKa = 3.65
MM1 pKa = 7.26ICIPRR6 pKa = 11.84KK7 pKa = 10.16AFLLCWSFLLLMVFVLLGILLGFFAQFRR35 pKa = 11.84KK36 pKa = 9.6FRR38 pKa = 11.84YY39 pKa = 9.49AGAKK43 pKa = 7.69VTFIPAATLPADD55 pKa = 4.06PLQVSYY61 pKa = 10.83EE62 pKa = 4.08AGEE65 pKa = 4.0PTIDD69 pKa = 5.06PRR71 pKa = 11.84DD72 pKa = 3.66LLNPILHH79 pKa = 6.5KK80 pKa = 10.66GIHH83 pKa = 6.41GDD85 pKa = 3.29ALGNLLEE92 pKa = 4.73EE93 pKa = 4.58YY94 pKa = 10.34KK95 pKa = 10.56QVLASGSSLEE105 pKa = 4.16LTKK108 pKa = 10.2TAAANYY114 pKa = 5.74MTLYY118 pKa = 8.82YY119 pKa = 10.33QCLTDD124 pKa = 4.1PSFKK128 pKa = 10.54KK129 pKa = 10.47SGLRR133 pKa = 11.84QGFVQNLYY141 pKa = 9.99PLVYY145 pKa = 10.58SLGTNRR151 pKa = 11.84QIMPSRR157 pKa = 11.84IFTAGASGLTVGDD170 pKa = 3.99PSLMEE175 pKa = 3.84TGKK178 pKa = 10.16IFSEE182 pKa = 4.09TDD184 pKa = 2.99GQFEE188 pKa = 4.8YY189 pKa = 11.17NDD191 pKa = 3.78EE192 pKa = 4.21TWGVSVRR199 pKa = 11.84RR200 pKa = 11.84SLGQAQQNLKK210 pKa = 10.96DD211 pKa = 4.51NAIEE215 pKa = 3.89WKK217 pKa = 9.37TYY219 pKa = 11.08DD220 pKa = 3.76EE221 pKa = 4.68FTTKK225 pKa = 9.01PTRR228 pKa = 11.84LGWLDD233 pKa = 3.31TLQIIRR239 pKa = 11.84DD240 pKa = 4.14NQSSMPDD247 pKa = 3.32SQVTGVNIEE256 pKa = 3.82LSIRR260 pKa = 11.84NPQDD264 pKa = 3.21PLASRR269 pKa = 11.84VTPCNFTTLPKK280 pKa = 10.03IPMYY284 pKa = 10.3MCFLPPAYY292 pKa = 8.44KK293 pKa = 9.61TEE295 pKa = 3.81MYY297 pKa = 10.14FRR299 pKa = 11.84IVIQHH304 pKa = 5.02YY305 pKa = 10.47FEE307 pKa = 4.84FKK309 pKa = 10.34EE310 pKa = 4.12FRR312 pKa = 11.84STFNQFGIGQVSTATDD328 pKa = 3.55VPYY331 pKa = 11.05DD332 pKa = 3.4WLNAIKK338 pKa = 10.12KK339 pKa = 9.68VSTASVEE346 pKa = 4.4SEE348 pKa = 4.08KK349 pKa = 11.27SLDD352 pKa = 3.46IAGGDD357 pKa = 4.0AEE359 pKa = 4.49LTTDD363 pKa = 3.97GVLL366 pKa = 3.65
Molecular weight: 40.99 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A076VEP0|A0A076VEP0_9VIRU Cap protein OS=Porcine associated porprismacovirus 9 OX=2170125 PE=4 SV=1
MM1 pKa = 8.06RR2 pKa = 11.84INAKK6 pKa = 8.49CTLVRR11 pKa = 11.84TIHH14 pKa = 6.67HH15 pKa = 7.33KK16 pKa = 9.8DD17 pKa = 2.81TLRR20 pKa = 11.84YY21 pKa = 10.37KK22 pKa = 10.12MVTNYY27 pKa = 9.76MITWSRR33 pKa = 11.84NDD35 pKa = 3.05TYY37 pKa = 11.53KK38 pKa = 10.67QFNQLMRR45 pKa = 11.84LIRR48 pKa = 11.84KK49 pKa = 9.19NDD51 pKa = 2.77IHH53 pKa = 7.31RR54 pKa = 11.84WIVGIEE60 pKa = 4.02TGTNGYY66 pKa = 9.6DD67 pKa = 2.99HH68 pKa = 6.68FQIRR72 pKa = 11.84MATRR76 pKa = 11.84WTFDD80 pKa = 2.72QMTEE84 pKa = 3.76IFHH87 pKa = 5.83GQAHH91 pKa = 6.37IEE93 pKa = 4.23EE94 pKa = 5.32CSDD97 pKa = 2.52TWEE100 pKa = 4.21YY101 pKa = 10.56EE102 pKa = 4.05RR103 pKa = 11.84KK104 pKa = 10.09SGIFFSSEE112 pKa = 3.21DD113 pKa = 3.37TAEE116 pKa = 3.67IRR118 pKa = 11.84RR119 pKa = 11.84VRR121 pKa = 11.84FGHH124 pKa = 6.45PNQAQEE130 pKa = 4.75HH131 pKa = 6.24IIEE134 pKa = 4.37TARR137 pKa = 11.84GQNDD141 pKa = 3.47RR142 pKa = 11.84QVDD145 pKa = 3.15VWYY148 pKa = 10.04DD149 pKa = 3.3PKK151 pKa = 11.36GNHH154 pKa = 5.59GKK156 pKa = 7.72TWLTVHH162 pKa = 5.87LWEE165 pKa = 5.31RR166 pKa = 11.84GQALVVPRR174 pKa = 11.84SSATPEE180 pKa = 3.89KK181 pKa = 10.46LSAFICSAYY190 pKa = 9.79KK191 pKa = 10.62GEE193 pKa = 4.17EE194 pKa = 4.13FIIIDD199 pKa = 3.62IPRR202 pKa = 11.84AGKK205 pKa = 9.94IPPSLYY211 pKa = 10.52EE212 pKa = 3.93VIEE215 pKa = 4.1EE216 pKa = 4.34VKK218 pKa = 10.79DD219 pKa = 3.6GLVFDD224 pKa = 4.47HH225 pKa = 7.49RR226 pKa = 11.84YY227 pKa = 7.42TGKK230 pKa = 8.17TRR232 pKa = 11.84NIRR235 pKa = 11.84GAKK238 pKa = 9.87VIIFTNTKK246 pKa = 10.09LDD248 pKa = 3.94EE249 pKa = 4.79KK250 pKa = 10.7KK251 pKa = 10.61LSYY254 pKa = 10.66DD255 pKa = 2.72RR256 pKa = 11.84WRR258 pKa = 11.84LHH260 pKa = 5.49GMEE263 pKa = 4.8KK264 pKa = 10.41GGEE267 pKa = 4.21GEE269 pKa = 4.12PLSS272 pKa = 4.04
MM1 pKa = 8.06RR2 pKa = 11.84INAKK6 pKa = 8.49CTLVRR11 pKa = 11.84TIHH14 pKa = 6.67HH15 pKa = 7.33KK16 pKa = 9.8DD17 pKa = 2.81TLRR20 pKa = 11.84YY21 pKa = 10.37KK22 pKa = 10.12MVTNYY27 pKa = 9.76MITWSRR33 pKa = 11.84NDD35 pKa = 3.05TYY37 pKa = 11.53KK38 pKa = 10.67QFNQLMRR45 pKa = 11.84LIRR48 pKa = 11.84KK49 pKa = 9.19NDD51 pKa = 2.77IHH53 pKa = 7.31RR54 pKa = 11.84WIVGIEE60 pKa = 4.02TGTNGYY66 pKa = 9.6DD67 pKa = 2.99HH68 pKa = 6.68FQIRR72 pKa = 11.84MATRR76 pKa = 11.84WTFDD80 pKa = 2.72QMTEE84 pKa = 3.76IFHH87 pKa = 5.83GQAHH91 pKa = 6.37IEE93 pKa = 4.23EE94 pKa = 5.32CSDD97 pKa = 2.52TWEE100 pKa = 4.21YY101 pKa = 10.56EE102 pKa = 4.05RR103 pKa = 11.84KK104 pKa = 10.09SGIFFSSEE112 pKa = 3.21DD113 pKa = 3.37TAEE116 pKa = 3.67IRR118 pKa = 11.84RR119 pKa = 11.84VRR121 pKa = 11.84FGHH124 pKa = 6.45PNQAQEE130 pKa = 4.75HH131 pKa = 6.24IIEE134 pKa = 4.37TARR137 pKa = 11.84GQNDD141 pKa = 3.47RR142 pKa = 11.84QVDD145 pKa = 3.15VWYY148 pKa = 10.04DD149 pKa = 3.3PKK151 pKa = 11.36GNHH154 pKa = 5.59GKK156 pKa = 7.72TWLTVHH162 pKa = 5.87LWEE165 pKa = 5.31RR166 pKa = 11.84GQALVVPRR174 pKa = 11.84SSATPEE180 pKa = 3.89KK181 pKa = 10.46LSAFICSAYY190 pKa = 9.79KK191 pKa = 10.62GEE193 pKa = 4.17EE194 pKa = 4.13FIIIDD199 pKa = 3.62IPRR202 pKa = 11.84AGKK205 pKa = 9.94IPPSLYY211 pKa = 10.52EE212 pKa = 3.93VIEE215 pKa = 4.1EE216 pKa = 4.34VKK218 pKa = 10.79DD219 pKa = 3.6GLVFDD224 pKa = 4.47HH225 pKa = 7.49RR226 pKa = 11.84YY227 pKa = 7.42TGKK230 pKa = 8.17TRR232 pKa = 11.84NIRR235 pKa = 11.84GAKK238 pKa = 9.87VIIFTNTKK246 pKa = 10.09LDD248 pKa = 3.94EE249 pKa = 4.79KK250 pKa = 10.7KK251 pKa = 10.61LSYY254 pKa = 10.66DD255 pKa = 2.72RR256 pKa = 11.84WRR258 pKa = 11.84LHH260 pKa = 5.49GMEE263 pKa = 4.8KK264 pKa = 10.41GGEE267 pKa = 4.21GEE269 pKa = 4.12PLSS272 pKa = 4.04
Molecular weight: 31.98 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
638 |
272 |
366 |
319.0 |
36.48 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
5.643 ± 0.781 | 1.254 ± 0.096 |
5.329 ± 0.118 | 6.113 ± 1.02 |
5.172 ± 0.716 | 6.897 ± 0.056 |
2.351 ± 1.307 | 7.053 ± 1.123 |
5.643 ± 0.618 | 8.621 ± 2.203 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.508 ± 0.042 | 3.918 ± 0.08 |
4.545 ± 1.017 | 4.389 ± 0.452 |
5.956 ± 1.585 | 5.799 ± 0.88 |
7.837 ± 0.074 | 5.016 ± 0.15 |
2.038 ± 0.573 | 3.918 ± 0.153 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
