
Genomoviridae sp.
Taxonomy: Viruses; Monodnaviria; Shotokuvirae; Cressdnaviricota; Repensiviricetes; Geplafuvirales; Genomoviridae; unclassified Genomoviridae
Average proteome isoelectric point is 6.76
Get precalculated fractions of proteins
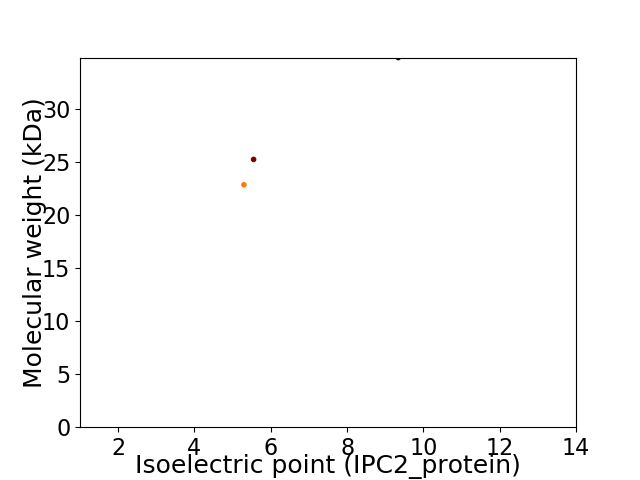
Virtual 2D-PAGE plot for 3 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
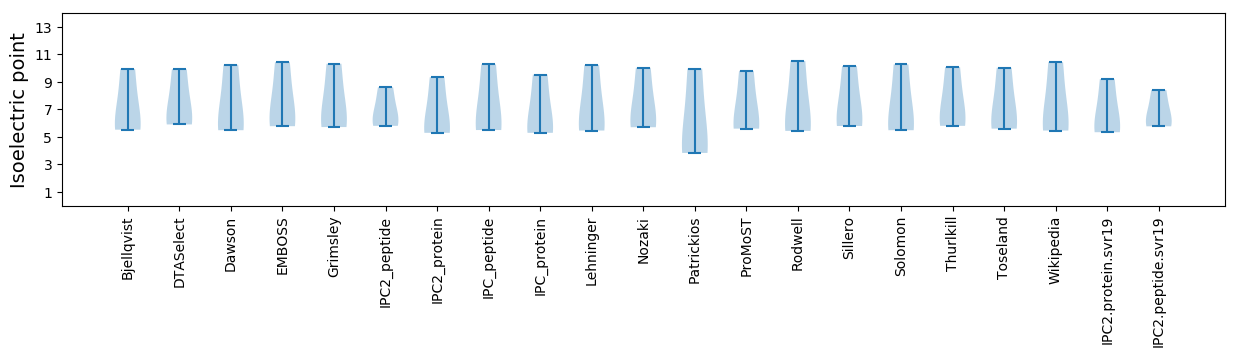
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A345N0J8|A0A345N0J8_9VIRU Rep catalytic domain protein OS=Genomoviridae sp. OX=2202565 PE=3 SV=1
MM1 pKa = 7.4TFDD4 pKa = 3.03FHH6 pKa = 7.8CRR8 pKa = 11.84YY9 pKa = 10.74GLFTYY14 pKa = 7.66SQCTGLDD21 pKa = 3.23PWHH24 pKa = 6.51VLDD27 pKa = 4.79KK28 pKa = 10.79FSEE31 pKa = 4.04IGAEE35 pKa = 4.04CIIARR40 pKa = 11.84EE41 pKa = 4.04AHH43 pKa = 6.55ADD45 pKa = 3.58GGTHH49 pKa = 6.19LHH51 pKa = 6.59VFADD55 pKa = 3.35WGRR58 pKa = 11.84RR59 pKa = 11.84KK60 pKa = 10.07RR61 pKa = 11.84FRR63 pKa = 11.84RR64 pKa = 11.84HH65 pKa = 6.15DD66 pKa = 3.67FADD69 pKa = 3.18VDD71 pKa = 4.21SFHH74 pKa = 7.12PNIVPSRR81 pKa = 11.84GTPHH85 pKa = 7.33LGWDD89 pKa = 3.69YY90 pKa = 10.71ATKK93 pKa = 10.79DD94 pKa = 3.25GDD96 pKa = 3.7IVAGGLEE103 pKa = 4.28RR104 pKa = 11.84PGGSCDD110 pKa = 3.06NLLRR114 pKa = 11.84KK115 pKa = 9.79DD116 pKa = 4.48HH117 pKa = 6.64IWNQIMDD124 pKa = 3.94AVTRR128 pKa = 11.84EE129 pKa = 3.85QFFEE133 pKa = 4.3LVRR136 pKa = 11.84EE137 pKa = 4.3LAPADD142 pKa = 3.69LAKK145 pKa = 10.71SFPSLSKK152 pKa = 10.52YY153 pKa = 10.8ADD155 pKa = 2.85WRR157 pKa = 11.84YY158 pKa = 10.63AEE160 pKa = 4.47PLATYY165 pKa = 8.2EE166 pKa = 4.51GPTFGDD172 pKa = 2.86IRR174 pKa = 11.84FDD176 pKa = 3.86LSPYY180 pKa = 9.73PGLEE184 pKa = 3.55QWCNDD189 pKa = 2.31IRR191 pKa = 11.84AGSNGQSGTYY201 pKa = 9.83
MM1 pKa = 7.4TFDD4 pKa = 3.03FHH6 pKa = 7.8CRR8 pKa = 11.84YY9 pKa = 10.74GLFTYY14 pKa = 7.66SQCTGLDD21 pKa = 3.23PWHH24 pKa = 6.51VLDD27 pKa = 4.79KK28 pKa = 10.79FSEE31 pKa = 4.04IGAEE35 pKa = 4.04CIIARR40 pKa = 11.84EE41 pKa = 4.04AHH43 pKa = 6.55ADD45 pKa = 3.58GGTHH49 pKa = 6.19LHH51 pKa = 6.59VFADD55 pKa = 3.35WGRR58 pKa = 11.84RR59 pKa = 11.84KK60 pKa = 10.07RR61 pKa = 11.84FRR63 pKa = 11.84RR64 pKa = 11.84HH65 pKa = 6.15DD66 pKa = 3.67FADD69 pKa = 3.18VDD71 pKa = 4.21SFHH74 pKa = 7.12PNIVPSRR81 pKa = 11.84GTPHH85 pKa = 7.33LGWDD89 pKa = 3.69YY90 pKa = 10.71ATKK93 pKa = 10.79DD94 pKa = 3.25GDD96 pKa = 3.7IVAGGLEE103 pKa = 4.28RR104 pKa = 11.84PGGSCDD110 pKa = 3.06NLLRR114 pKa = 11.84KK115 pKa = 9.79DD116 pKa = 4.48HH117 pKa = 6.64IWNQIMDD124 pKa = 3.94AVTRR128 pKa = 11.84EE129 pKa = 3.85QFFEE133 pKa = 4.3LVRR136 pKa = 11.84EE137 pKa = 4.3LAPADD142 pKa = 3.69LAKK145 pKa = 10.71SFPSLSKK152 pKa = 10.52YY153 pKa = 10.8ADD155 pKa = 2.85WRR157 pKa = 11.84YY158 pKa = 10.63AEE160 pKa = 4.47PLATYY165 pKa = 8.2EE166 pKa = 4.51GPTFGDD172 pKa = 2.86IRR174 pKa = 11.84FDD176 pKa = 3.86LSPYY180 pKa = 9.73PGLEE184 pKa = 3.55QWCNDD189 pKa = 2.31IRR191 pKa = 11.84AGSNGQSGTYY201 pKa = 9.83
Molecular weight: 22.84 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A345N0J7|A0A345N0J7_9VIRU Uncharacterized protein OS=Genomoviridae sp. OX=2202565 PE=4 SV=1
MM1 pKa = 6.37ATKK4 pKa = 10.45RR5 pKa = 11.84IIRR8 pKa = 11.84PRR10 pKa = 11.84RR11 pKa = 11.84GNRR14 pKa = 11.84RR15 pKa = 11.84FVKK18 pKa = 10.43RR19 pKa = 11.84KK20 pKa = 8.89ARR22 pKa = 11.84PAARR26 pKa = 11.84SRR28 pKa = 11.84RR29 pKa = 11.84RR30 pKa = 11.84PMKK33 pKa = 9.92RR34 pKa = 11.84GSMSRR39 pKa = 11.84KK40 pKa = 9.05RR41 pKa = 11.84ILNITSRR48 pKa = 11.84KK49 pKa = 8.82KK50 pKa = 9.49QDD52 pKa = 4.09NMRR55 pKa = 11.84SWNGTPFLAGSAGPIVMQGDD75 pKa = 4.0DD76 pKa = 3.51SYY78 pKa = 11.72MFGFVCTARR87 pKa = 11.84DD88 pKa = 3.52KK89 pKa = 11.58DD90 pKa = 4.33LGTGPAPIEE99 pKa = 4.01MDD101 pKa = 3.61PSRR104 pKa = 11.84TSTHH108 pKa = 6.39CYY110 pKa = 8.37MRR112 pKa = 11.84GFKK115 pKa = 10.29EE116 pKa = 4.69RR117 pKa = 11.84IDD119 pKa = 4.22LFTGSAHH126 pKa = 5.37PWIWRR131 pKa = 11.84RR132 pKa = 11.84ILFTYY137 pKa = 10.06KK138 pKa = 10.69GLDD141 pKa = 3.63LLRR144 pKa = 11.84VTDD147 pKa = 4.87DD148 pKa = 4.15EE149 pKa = 5.12SPGDD153 pKa = 4.04DD154 pKa = 3.68GVGQLWVEE162 pKa = 4.11SSDD165 pKa = 3.81GYY167 pKa = 10.74GRR169 pKa = 11.84YY170 pKa = 8.74YY171 pKa = 11.4ANIGDD176 pKa = 4.01PTDD179 pKa = 4.04LKK181 pKa = 11.25SSVIGQHH188 pKa = 5.84IRR190 pKa = 11.84DD191 pKa = 4.7LIFKK195 pKa = 8.17GTAEE199 pKa = 4.19KK200 pKa = 10.52DD201 pKa = 3.61YY202 pKa = 10.85TSSFSAKK209 pKa = 9.1TDD211 pKa = 3.36QEE213 pKa = 4.33RR214 pKa = 11.84VKK216 pKa = 10.47ILYY219 pKa = 9.93DD220 pKa = 3.12KK221 pKa = 10.79TVTIKK226 pKa = 10.55SGNDD230 pKa = 2.48RR231 pKa = 11.84GVIKK235 pKa = 9.44TYY237 pKa = 10.62RR238 pKa = 11.84RR239 pKa = 11.84WHH241 pKa = 5.65GFNRR245 pKa = 11.84TLIYY249 pKa = 11.03DD250 pKa = 3.75EE251 pKa = 5.68DD252 pKa = 4.0EE253 pKa = 4.62VGAQTANRR261 pKa = 11.84LLSSQSPRR269 pKa = 11.84SMGDD273 pKa = 3.0VYY275 pKa = 11.43VIDD278 pKa = 4.77FFSPGLGGTSDD289 pKa = 3.97DD290 pKa = 4.35EE291 pKa = 4.97LRR293 pKa = 11.84LLPTATLYY301 pKa = 8.93WHH303 pKa = 6.59EE304 pKa = 4.22RR305 pKa = 3.47
MM1 pKa = 6.37ATKK4 pKa = 10.45RR5 pKa = 11.84IIRR8 pKa = 11.84PRR10 pKa = 11.84RR11 pKa = 11.84GNRR14 pKa = 11.84RR15 pKa = 11.84FVKK18 pKa = 10.43RR19 pKa = 11.84KK20 pKa = 8.89ARR22 pKa = 11.84PAARR26 pKa = 11.84SRR28 pKa = 11.84RR29 pKa = 11.84RR30 pKa = 11.84PMKK33 pKa = 9.92RR34 pKa = 11.84GSMSRR39 pKa = 11.84KK40 pKa = 9.05RR41 pKa = 11.84ILNITSRR48 pKa = 11.84KK49 pKa = 8.82KK50 pKa = 9.49QDD52 pKa = 4.09NMRR55 pKa = 11.84SWNGTPFLAGSAGPIVMQGDD75 pKa = 4.0DD76 pKa = 3.51SYY78 pKa = 11.72MFGFVCTARR87 pKa = 11.84DD88 pKa = 3.52KK89 pKa = 11.58DD90 pKa = 4.33LGTGPAPIEE99 pKa = 4.01MDD101 pKa = 3.61PSRR104 pKa = 11.84TSTHH108 pKa = 6.39CYY110 pKa = 8.37MRR112 pKa = 11.84GFKK115 pKa = 10.29EE116 pKa = 4.69RR117 pKa = 11.84IDD119 pKa = 4.22LFTGSAHH126 pKa = 5.37PWIWRR131 pKa = 11.84RR132 pKa = 11.84ILFTYY137 pKa = 10.06KK138 pKa = 10.69GLDD141 pKa = 3.63LLRR144 pKa = 11.84VTDD147 pKa = 4.87DD148 pKa = 4.15EE149 pKa = 5.12SPGDD153 pKa = 4.04DD154 pKa = 3.68GVGQLWVEE162 pKa = 4.11SSDD165 pKa = 3.81GYY167 pKa = 10.74GRR169 pKa = 11.84YY170 pKa = 8.74YY171 pKa = 11.4ANIGDD176 pKa = 4.01PTDD179 pKa = 4.04LKK181 pKa = 11.25SSVIGQHH188 pKa = 5.84IRR190 pKa = 11.84DD191 pKa = 4.7LIFKK195 pKa = 8.17GTAEE199 pKa = 4.19KK200 pKa = 10.52DD201 pKa = 3.61YY202 pKa = 10.85TSSFSAKK209 pKa = 9.1TDD211 pKa = 3.36QEE213 pKa = 4.33RR214 pKa = 11.84VKK216 pKa = 10.47ILYY219 pKa = 9.93DD220 pKa = 3.12KK221 pKa = 10.79TVTIKK226 pKa = 10.55SGNDD230 pKa = 2.48RR231 pKa = 11.84GVIKK235 pKa = 9.44TYY237 pKa = 10.62RR238 pKa = 11.84RR239 pKa = 11.84WHH241 pKa = 5.65GFNRR245 pKa = 11.84TLIYY249 pKa = 11.03DD250 pKa = 3.75EE251 pKa = 5.68DD252 pKa = 4.0EE253 pKa = 4.62VGAQTANRR261 pKa = 11.84LLSSQSPRR269 pKa = 11.84SMGDD273 pKa = 3.0VYY275 pKa = 11.43VIDD278 pKa = 4.77FFSPGLGGTSDD289 pKa = 3.97DD290 pKa = 4.35EE291 pKa = 4.97LRR293 pKa = 11.84LLPTATLYY301 pKa = 8.93WHH303 pKa = 6.59EE304 pKa = 4.22RR305 pKa = 3.47
Molecular weight: 34.81 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
738 |
201 |
305 |
246.0 |
27.62 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.504 ± 0.728 | 1.355 ± 0.381 |
7.317 ± 1.395 | 4.065 ± 0.371 |
4.472 ± 0.655 | 8.537 ± 0.663 |
3.252 ± 0.744 | 5.556 ± 0.347 |
3.659 ± 1.155 | 9.485 ± 2.282 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
3.252 ± 1.018 | 2.71 ± 0.126 |
4.336 ± 0.556 | 3.117 ± 0.66 |
8.13 ± 1.788 | 9.485 ± 2.142 |
6.098 ± 0.515 | 3.794 ± 0.219 |
2.168 ± 0.271 | 2.71 ± 1.096 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
