
Talpa europaea papillomavirus 1
Taxonomy: Viruses; Monodnaviria; Shotokuvirae; Cossaviricota; Papovaviricetes; Zurhausenvirales; Papillomaviridae; Firstpapillomavirinae; Dyophipapillomavirus; Dyophipapillomavirus 1
Average proteome isoelectric point is 6.1
Get precalculated fractions of proteins
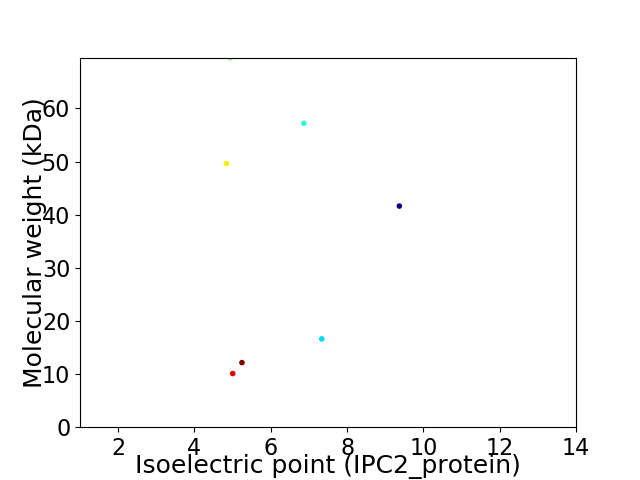
Virtual 2D-PAGE plot for 7 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|R9RYS7|R9RYS7_9PAPI Minor capsid protein L2 OS=Talpa europaea papillomavirus 1 OX=1338506 GN=L2 PE=3 SV=1
MM1 pKa = 7.65IGPEE5 pKa = 4.18PTLKK9 pKa = 10.63DD10 pKa = 3.2ITLDD14 pKa = 3.67TPDD17 pKa = 5.43SIDD20 pKa = 3.63LNCYY24 pKa = 6.56EE25 pKa = 4.06TFEE28 pKa = 4.3IEE30 pKa = 4.25EE31 pKa = 4.46EE32 pKa = 4.18EE33 pKa = 4.45VPPLGQPIKK42 pKa = 9.99VLCYY46 pKa = 10.24CGLCKK51 pKa = 10.06RR52 pKa = 11.84RR53 pKa = 11.84IKK55 pKa = 9.8VTVVSDD61 pKa = 3.65RR62 pKa = 11.84ASLISLEE69 pKa = 4.07SLLVDD74 pKa = 3.52SSLRR78 pKa = 11.84FVCCSCGRR86 pKa = 11.84RR87 pKa = 11.84YY88 pKa = 10.45GRR90 pKa = 4.21
MM1 pKa = 7.65IGPEE5 pKa = 4.18PTLKK9 pKa = 10.63DD10 pKa = 3.2ITLDD14 pKa = 3.67TPDD17 pKa = 5.43SIDD20 pKa = 3.63LNCYY24 pKa = 6.56EE25 pKa = 4.06TFEE28 pKa = 4.3IEE30 pKa = 4.25EE31 pKa = 4.46EE32 pKa = 4.18EE33 pKa = 4.45VPPLGQPIKK42 pKa = 9.99VLCYY46 pKa = 10.24CGLCKK51 pKa = 10.06RR52 pKa = 11.84RR53 pKa = 11.84IKK55 pKa = 9.8VTVVSDD61 pKa = 3.65RR62 pKa = 11.84ASLISLEE69 pKa = 4.07SLLVDD74 pKa = 3.52SSLRR78 pKa = 11.84FVCCSCGRR86 pKa = 11.84RR87 pKa = 11.84YY88 pKa = 10.45GRR90 pKa = 4.21
Molecular weight: 10.1 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|R9RZM2|R9RZM2_9PAPI Regulatory protein E2 OS=Talpa europaea papillomavirus 1 OX=1338506 GN=E2 PE=3 SV=1
MM1 pKa = 7.14EE2 pKa = 4.63TLKK5 pKa = 10.56QRR7 pKa = 11.84LDD9 pKa = 3.44SLQNEE14 pKa = 4.42LLSLYY19 pKa = 10.33EE20 pKa = 4.53KK21 pKa = 10.56DD22 pKa = 3.79SNDD25 pKa = 2.84IRR27 pKa = 11.84DD28 pKa = 4.87QILHH32 pKa = 6.0WNLNRR37 pKa = 11.84RR38 pKa = 11.84EE39 pKa = 4.03QALLHH44 pKa = 5.75YY45 pKa = 10.38ARR47 pKa = 11.84QQGLNRR53 pKa = 11.84LGMQMVPALTVSQSRR68 pKa = 11.84AKK70 pKa = 10.58GAIEE74 pKa = 4.15MEE76 pKa = 4.27LMLTSLSTSRR86 pKa = 11.84FGNEE90 pKa = 2.99GWTLQEE96 pKa = 3.87TSRR99 pKa = 11.84EE100 pKa = 4.18LFNTAPKK107 pKa = 9.0HH108 pKa = 4.5TFKK111 pKa = 10.84KK112 pKa = 10.42GPRR115 pKa = 11.84HH116 pKa = 6.45LDD118 pKa = 3.19MIFDD122 pKa = 4.28GNSNNTVHH130 pKa = 6.26VPFWQYY136 pKa = 10.73IYY138 pKa = 10.89YY139 pKa = 9.86QDD141 pKa = 5.49EE142 pKa = 3.78RR143 pKa = 11.84DD144 pKa = 3.12EE145 pKa = 4.22WHH147 pKa = 7.02RR148 pKa = 11.84SNSDD152 pKa = 2.85VDD154 pKa = 4.07SVGIYY159 pKa = 10.52YY160 pKa = 8.27MQGTIKK166 pKa = 10.5VYY168 pKa = 7.4YY169 pKa = 10.05TKK171 pKa = 10.57FQSEE175 pKa = 3.77ASKK178 pKa = 11.14YY179 pKa = 10.38GFRR182 pKa = 11.84TWQVSHH188 pKa = 7.38DD189 pKa = 4.07NKK191 pKa = 10.06PLHH194 pKa = 6.68SSSSVGSRR202 pKa = 11.84GPSPHH207 pKa = 6.69SKK209 pKa = 10.68APSTPIRR216 pKa = 11.84RR217 pKa = 11.84GGGRR221 pKa = 11.84RR222 pKa = 11.84APRR225 pKa = 11.84RR226 pKa = 11.84RR227 pKa = 11.84SPSTFSRR234 pKa = 11.84SHH236 pKa = 6.69KK237 pKa = 9.39EE238 pKa = 3.6ASGSSGRR245 pKa = 11.84GEE247 pKa = 4.43TPPSARR253 pKa = 11.84DD254 pKa = 3.32VGRR257 pKa = 11.84STEE260 pKa = 4.04TPPRR264 pKa = 11.84GNQSRR269 pKa = 11.84LRR271 pKa = 11.84SLLHH275 pKa = 6.17EE276 pKa = 4.76ARR278 pKa = 11.84DD279 pKa = 3.88PPLLVLKK286 pKa = 10.9GPANTLKK293 pKa = 10.6CLRR296 pKa = 11.84YY297 pKa = 8.76RR298 pKa = 11.84LKK300 pKa = 11.11GRR302 pKa = 11.84FSHH305 pKa = 6.22TFCNVTSTWHH315 pKa = 4.85WTLPTGTEE323 pKa = 3.86KK324 pKa = 10.66CVRR327 pKa = 11.84SCGSRR332 pKa = 11.84MIVTFSSTEE341 pKa = 3.7QRR343 pKa = 11.84NLFCTKK349 pKa = 10.01VPLPSSVTAFTGSFNGEE366 pKa = 3.63
MM1 pKa = 7.14EE2 pKa = 4.63TLKK5 pKa = 10.56QRR7 pKa = 11.84LDD9 pKa = 3.44SLQNEE14 pKa = 4.42LLSLYY19 pKa = 10.33EE20 pKa = 4.53KK21 pKa = 10.56DD22 pKa = 3.79SNDD25 pKa = 2.84IRR27 pKa = 11.84DD28 pKa = 4.87QILHH32 pKa = 6.0WNLNRR37 pKa = 11.84RR38 pKa = 11.84EE39 pKa = 4.03QALLHH44 pKa = 5.75YY45 pKa = 10.38ARR47 pKa = 11.84QQGLNRR53 pKa = 11.84LGMQMVPALTVSQSRR68 pKa = 11.84AKK70 pKa = 10.58GAIEE74 pKa = 4.15MEE76 pKa = 4.27LMLTSLSTSRR86 pKa = 11.84FGNEE90 pKa = 2.99GWTLQEE96 pKa = 3.87TSRR99 pKa = 11.84EE100 pKa = 4.18LFNTAPKK107 pKa = 9.0HH108 pKa = 4.5TFKK111 pKa = 10.84KK112 pKa = 10.42GPRR115 pKa = 11.84HH116 pKa = 6.45LDD118 pKa = 3.19MIFDD122 pKa = 4.28GNSNNTVHH130 pKa = 6.26VPFWQYY136 pKa = 10.73IYY138 pKa = 10.89YY139 pKa = 9.86QDD141 pKa = 5.49EE142 pKa = 3.78RR143 pKa = 11.84DD144 pKa = 3.12EE145 pKa = 4.22WHH147 pKa = 7.02RR148 pKa = 11.84SNSDD152 pKa = 2.85VDD154 pKa = 4.07SVGIYY159 pKa = 10.52YY160 pKa = 8.27MQGTIKK166 pKa = 10.5VYY168 pKa = 7.4YY169 pKa = 10.05TKK171 pKa = 10.57FQSEE175 pKa = 3.77ASKK178 pKa = 11.14YY179 pKa = 10.38GFRR182 pKa = 11.84TWQVSHH188 pKa = 7.38DD189 pKa = 4.07NKK191 pKa = 10.06PLHH194 pKa = 6.68SSSSVGSRR202 pKa = 11.84GPSPHH207 pKa = 6.69SKK209 pKa = 10.68APSTPIRR216 pKa = 11.84RR217 pKa = 11.84GGGRR221 pKa = 11.84RR222 pKa = 11.84APRR225 pKa = 11.84RR226 pKa = 11.84RR227 pKa = 11.84SPSTFSRR234 pKa = 11.84SHH236 pKa = 6.69KK237 pKa = 9.39EE238 pKa = 3.6ASGSSGRR245 pKa = 11.84GEE247 pKa = 4.43TPPSARR253 pKa = 11.84DD254 pKa = 3.32VGRR257 pKa = 11.84STEE260 pKa = 4.04TPPRR264 pKa = 11.84GNQSRR269 pKa = 11.84LRR271 pKa = 11.84SLLHH275 pKa = 6.17EE276 pKa = 4.76ARR278 pKa = 11.84DD279 pKa = 3.88PPLLVLKK286 pKa = 10.9GPANTLKK293 pKa = 10.6CLRR296 pKa = 11.84YY297 pKa = 8.76RR298 pKa = 11.84LKK300 pKa = 11.11GRR302 pKa = 11.84FSHH305 pKa = 6.22TFCNVTSTWHH315 pKa = 4.85WTLPTGTEE323 pKa = 3.86KK324 pKa = 10.66CVRR327 pKa = 11.84SCGSRR332 pKa = 11.84MIVTFSSTEE341 pKa = 3.7QRR343 pKa = 11.84NLFCTKK349 pKa = 10.01VPLPSSVTAFTGSFNGEE366 pKa = 3.63
Molecular weight: 41.62 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
2265 |
90 |
608 |
323.6 |
36.69 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
4.901 ± 0.575 | 1.987 ± 0.548 |
5.563 ± 0.576 | 7.285 ± 0.591 |
4.812 ± 0.443 | 5.916 ± 0.823 |
1.987 ± 0.332 | 4.989 ± 0.745 |
5.21 ± 0.825 | 9.36 ± 0.844 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.943 ± 0.458 | 4.459 ± 0.717 |
5.916 ± 1.017 | 4.194 ± 0.407 |
6.446 ± 0.885 | 8.079 ± 1.282 |
6.313 ± 0.569 | 6.269 ± 0.582 |
1.369 ± 0.304 | 3.002 ± 0.265 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
