
Bacillus alcalophilus ATCC 27647 = CGMCC 1.3604
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Firmicutes; Bacilli; Bacillales; Bacillaceae; Alkalihalobacillus; Alkalihalobacillus alcalophilus
Average proteome isoelectric point is 6.2
Get precalculated fractions of proteins
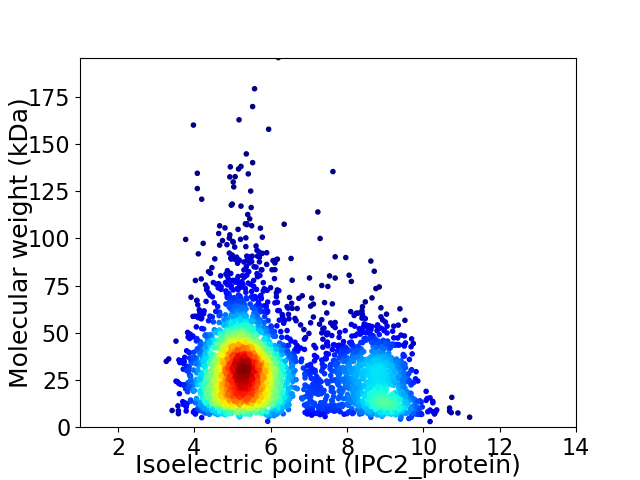
Virtual 2D-PAGE plot for 3743 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|J8T7M4|J8T7M4_ALKAL Iron transporter OS=Bacillus alcalophilus ATCC 27647 = CGMCC 1.3604 OX=1218173 GN=AJ85_03855 PE=3 SV=1
MM1 pKa = 7.22NNKK4 pKa = 8.94LRR6 pKa = 11.84LLLLVLLASFVVFMAACSNDD26 pKa = 3.51GASGDD31 pKa = 3.99PEE33 pKa = 4.42EE34 pKa = 5.06TPSDD38 pKa = 3.7DD39 pKa = 4.23PEE41 pKa = 4.32TEE43 pKa = 4.02EE44 pKa = 4.27TPEE47 pKa = 4.1EE48 pKa = 4.31DD49 pKa = 3.81EE50 pKa = 4.54EE51 pKa = 4.54TDD53 pKa = 3.26GLYY56 pKa = 10.66SIEE59 pKa = 4.91DD60 pKa = 3.5FDD62 pKa = 5.8HH63 pKa = 6.95IKK65 pKa = 11.05DD66 pKa = 4.31DD67 pKa = 3.59GTTIDD72 pKa = 3.49GGEE75 pKa = 4.16LVFGLSSDD83 pKa = 3.93TPFEE87 pKa = 4.31GTLNWNFYY95 pKa = 10.58SGNPDD100 pKa = 3.18AQVLQWFDD108 pKa = 3.83EE109 pKa = 4.52PLLTWDD115 pKa = 3.48EE116 pKa = 4.56NYY118 pKa = 10.6VYY120 pKa = 9.5TQDD123 pKa = 3.41GAATYY128 pKa = 10.16EE129 pKa = 3.97IDD131 pKa = 4.82EE132 pKa = 4.5EE133 pKa = 4.77DD134 pKa = 3.46NRR136 pKa = 11.84IWTLTIRR143 pKa = 11.84DD144 pKa = 3.82NVNWHH149 pKa = 7.33DD150 pKa = 4.04GTPVTAEE157 pKa = 3.7DD158 pKa = 3.32WAFAHH163 pKa = 6.28YY164 pKa = 10.42VIADD168 pKa = 3.62PDD170 pKa = 3.91YY171 pKa = 11.31DD172 pKa = 3.65GVRR175 pKa = 11.84FDD177 pKa = 3.26SSLRR181 pKa = 11.84NIEE184 pKa = 4.67GIVDD188 pKa = 3.67YY189 pKa = 11.29HH190 pKa = 7.95NGEE193 pKa = 4.02ADD195 pKa = 4.02EE196 pKa = 4.48ISGIEE201 pKa = 4.17IIDD204 pKa = 3.84EE205 pKa = 3.9KK206 pKa = 9.65TLRR209 pKa = 11.84ITYY212 pKa = 8.83VQATPSLITGGVWTYY227 pKa = 11.04PLAKK231 pKa = 9.53HH232 pKa = 6.49IYY234 pKa = 8.65GDD236 pKa = 3.63IPVEE240 pKa = 4.15EE241 pKa = 4.56MSSHH245 pKa = 6.61AATRR249 pKa = 11.84QNPIGFGPYY258 pKa = 9.53KK259 pKa = 10.14VDD261 pKa = 3.7SIVPGEE267 pKa = 4.1SVSYY271 pKa = 10.57SKK273 pKa = 11.65YY274 pKa = 7.66EE275 pKa = 4.06DD276 pKa = 3.52YY277 pKa = 10.71WRR279 pKa = 11.84GEE281 pKa = 3.8PALDD285 pKa = 3.33KK286 pKa = 10.56VTLRR290 pKa = 11.84VINPSVVVEE299 pKa = 3.91EE300 pKa = 4.37LRR302 pKa = 11.84NGGVDD307 pKa = 3.39MVSSFPQAQFADD319 pKa = 4.12VVDD322 pKa = 4.17EE323 pKa = 4.59LSNITWLGNIDD334 pKa = 3.59RR335 pKa = 11.84AYY337 pKa = 9.76TYY339 pKa = 10.47IGFKK343 pKa = 10.12LGHH346 pKa = 6.36WDD348 pKa = 3.13EE349 pKa = 4.37EE350 pKa = 4.41NRR352 pKa = 11.84VNVYY356 pKa = 10.56DD357 pKa = 4.15PEE359 pKa = 4.11NAKK362 pKa = 9.68MGNKK366 pKa = 9.27NLRR369 pKa = 11.84QAMWHH374 pKa = 6.19AVDD377 pKa = 4.01NDD379 pKa = 4.18TVGEE383 pKa = 4.07RR384 pKa = 11.84FYY386 pKa = 11.27HH387 pKa = 5.77GLRR390 pKa = 11.84WAGTTLIPPSHH401 pKa = 7.25PEE403 pKa = 3.53FHH405 pKa = 7.6DD406 pKa = 3.32ATNPGRR412 pKa = 11.84AYY414 pKa = 10.58DD415 pKa = 3.76PEE417 pKa = 4.3RR418 pKa = 11.84ANEE421 pKa = 4.09LLDD424 pKa = 3.41EE425 pKa = 4.96AGYY428 pKa = 11.28VDD430 pKa = 3.81VDD432 pKa = 3.24GDD434 pKa = 4.08GFRR437 pKa = 11.84EE438 pKa = 4.26DD439 pKa = 3.74PDD441 pKa = 4.02GNEE444 pKa = 3.83LVINFASMSGDD455 pKa = 3.64EE456 pKa = 4.22IAEE459 pKa = 4.06PLANYY464 pKa = 8.64YY465 pKa = 8.96IQAWANVGLNVQLLDD480 pKa = 3.91GRR482 pKa = 11.84LHH484 pKa = 6.49EE485 pKa = 5.13FNSFYY490 pKa = 11.31DD491 pKa = 3.47RR492 pKa = 11.84VEE494 pKa = 4.91ADD496 pKa = 4.8DD497 pKa = 4.81PDD499 pKa = 4.01IDD501 pKa = 3.81IYY503 pKa = 10.86QGAWVVGIDD512 pKa = 3.62VDD514 pKa = 4.23PSGLYY519 pKa = 10.12GAEE522 pKa = 3.24ASYY525 pKa = 11.38NYY527 pKa = 8.89TRR529 pKa = 11.84YY530 pKa = 10.52VSDD533 pKa = 4.16EE534 pKa = 3.94NNQLMTDD541 pKa = 4.32GLSEE545 pKa = 3.95AAFDD549 pKa = 3.04VDD551 pKa = 3.95YY552 pKa = 11.13RR553 pKa = 11.84VDD555 pKa = 5.23VYY557 pKa = 11.14NQWQEE562 pKa = 3.92YY563 pKa = 7.28MVEE566 pKa = 4.18NVPAFPTLYY575 pKa = 10.03RR576 pKa = 11.84AALVPVNNRR585 pKa = 11.84VTGYY589 pKa = 11.13AVGDD593 pKa = 3.44GTGYY597 pKa = 10.15FRR599 pKa = 11.84YY600 pKa = 9.42QVGVTEE606 pKa = 4.67EE607 pKa = 4.12VAEE610 pKa = 4.23SAEE613 pKa = 4.02
MM1 pKa = 7.22NNKK4 pKa = 8.94LRR6 pKa = 11.84LLLLVLLASFVVFMAACSNDD26 pKa = 3.51GASGDD31 pKa = 3.99PEE33 pKa = 4.42EE34 pKa = 5.06TPSDD38 pKa = 3.7DD39 pKa = 4.23PEE41 pKa = 4.32TEE43 pKa = 4.02EE44 pKa = 4.27TPEE47 pKa = 4.1EE48 pKa = 4.31DD49 pKa = 3.81EE50 pKa = 4.54EE51 pKa = 4.54TDD53 pKa = 3.26GLYY56 pKa = 10.66SIEE59 pKa = 4.91DD60 pKa = 3.5FDD62 pKa = 5.8HH63 pKa = 6.95IKK65 pKa = 11.05DD66 pKa = 4.31DD67 pKa = 3.59GTTIDD72 pKa = 3.49GGEE75 pKa = 4.16LVFGLSSDD83 pKa = 3.93TPFEE87 pKa = 4.31GTLNWNFYY95 pKa = 10.58SGNPDD100 pKa = 3.18AQVLQWFDD108 pKa = 3.83EE109 pKa = 4.52PLLTWDD115 pKa = 3.48EE116 pKa = 4.56NYY118 pKa = 10.6VYY120 pKa = 9.5TQDD123 pKa = 3.41GAATYY128 pKa = 10.16EE129 pKa = 3.97IDD131 pKa = 4.82EE132 pKa = 4.5EE133 pKa = 4.77DD134 pKa = 3.46NRR136 pKa = 11.84IWTLTIRR143 pKa = 11.84DD144 pKa = 3.82NVNWHH149 pKa = 7.33DD150 pKa = 4.04GTPVTAEE157 pKa = 3.7DD158 pKa = 3.32WAFAHH163 pKa = 6.28YY164 pKa = 10.42VIADD168 pKa = 3.62PDD170 pKa = 3.91YY171 pKa = 11.31DD172 pKa = 3.65GVRR175 pKa = 11.84FDD177 pKa = 3.26SSLRR181 pKa = 11.84NIEE184 pKa = 4.67GIVDD188 pKa = 3.67YY189 pKa = 11.29HH190 pKa = 7.95NGEE193 pKa = 4.02ADD195 pKa = 4.02EE196 pKa = 4.48ISGIEE201 pKa = 4.17IIDD204 pKa = 3.84EE205 pKa = 3.9KK206 pKa = 9.65TLRR209 pKa = 11.84ITYY212 pKa = 8.83VQATPSLITGGVWTYY227 pKa = 11.04PLAKK231 pKa = 9.53HH232 pKa = 6.49IYY234 pKa = 8.65GDD236 pKa = 3.63IPVEE240 pKa = 4.15EE241 pKa = 4.56MSSHH245 pKa = 6.61AATRR249 pKa = 11.84QNPIGFGPYY258 pKa = 9.53KK259 pKa = 10.14VDD261 pKa = 3.7SIVPGEE267 pKa = 4.1SVSYY271 pKa = 10.57SKK273 pKa = 11.65YY274 pKa = 7.66EE275 pKa = 4.06DD276 pKa = 3.52YY277 pKa = 10.71WRR279 pKa = 11.84GEE281 pKa = 3.8PALDD285 pKa = 3.33KK286 pKa = 10.56VTLRR290 pKa = 11.84VINPSVVVEE299 pKa = 3.91EE300 pKa = 4.37LRR302 pKa = 11.84NGGVDD307 pKa = 3.39MVSSFPQAQFADD319 pKa = 4.12VVDD322 pKa = 4.17EE323 pKa = 4.59LSNITWLGNIDD334 pKa = 3.59RR335 pKa = 11.84AYY337 pKa = 9.76TYY339 pKa = 10.47IGFKK343 pKa = 10.12LGHH346 pKa = 6.36WDD348 pKa = 3.13EE349 pKa = 4.37EE350 pKa = 4.41NRR352 pKa = 11.84VNVYY356 pKa = 10.56DD357 pKa = 4.15PEE359 pKa = 4.11NAKK362 pKa = 9.68MGNKK366 pKa = 9.27NLRR369 pKa = 11.84QAMWHH374 pKa = 6.19AVDD377 pKa = 4.01NDD379 pKa = 4.18TVGEE383 pKa = 4.07RR384 pKa = 11.84FYY386 pKa = 11.27HH387 pKa = 5.77GLRR390 pKa = 11.84WAGTTLIPPSHH401 pKa = 7.25PEE403 pKa = 3.53FHH405 pKa = 7.6DD406 pKa = 3.32ATNPGRR412 pKa = 11.84AYY414 pKa = 10.58DD415 pKa = 3.76PEE417 pKa = 4.3RR418 pKa = 11.84ANEE421 pKa = 4.09LLDD424 pKa = 3.41EE425 pKa = 4.96AGYY428 pKa = 11.28VDD430 pKa = 3.81VDD432 pKa = 3.24GDD434 pKa = 4.08GFRR437 pKa = 11.84EE438 pKa = 4.26DD439 pKa = 3.74PDD441 pKa = 4.02GNEE444 pKa = 3.83LVINFASMSGDD455 pKa = 3.64EE456 pKa = 4.22IAEE459 pKa = 4.06PLANYY464 pKa = 8.64YY465 pKa = 8.96IQAWANVGLNVQLLDD480 pKa = 3.91GRR482 pKa = 11.84LHH484 pKa = 6.49EE485 pKa = 5.13FNSFYY490 pKa = 11.31DD491 pKa = 3.47RR492 pKa = 11.84VEE494 pKa = 4.91ADD496 pKa = 4.8DD497 pKa = 4.81PDD499 pKa = 4.01IDD501 pKa = 3.81IYY503 pKa = 10.86QGAWVVGIDD512 pKa = 3.62VDD514 pKa = 4.23PSGLYY519 pKa = 10.12GAEE522 pKa = 3.24ASYY525 pKa = 11.38NYY527 pKa = 8.89TRR529 pKa = 11.84YY530 pKa = 10.52VSDD533 pKa = 4.16EE534 pKa = 3.94NNQLMTDD541 pKa = 4.32GLSEE545 pKa = 3.95AAFDD549 pKa = 3.04VDD551 pKa = 3.95YY552 pKa = 11.13RR553 pKa = 11.84VDD555 pKa = 5.23VYY557 pKa = 11.14NQWQEE562 pKa = 3.92YY563 pKa = 7.28MVEE566 pKa = 4.18NVPAFPTLYY575 pKa = 10.03RR576 pKa = 11.84AALVPVNNRR585 pKa = 11.84VTGYY589 pKa = 11.13AVGDD593 pKa = 3.44GTGYY597 pKa = 10.15FRR599 pKa = 11.84YY600 pKa = 9.42QVGVTEE606 pKa = 4.67EE607 pKa = 4.12VAEE610 pKa = 4.23SAEE613 pKa = 4.02
Molecular weight: 68.85 kDa
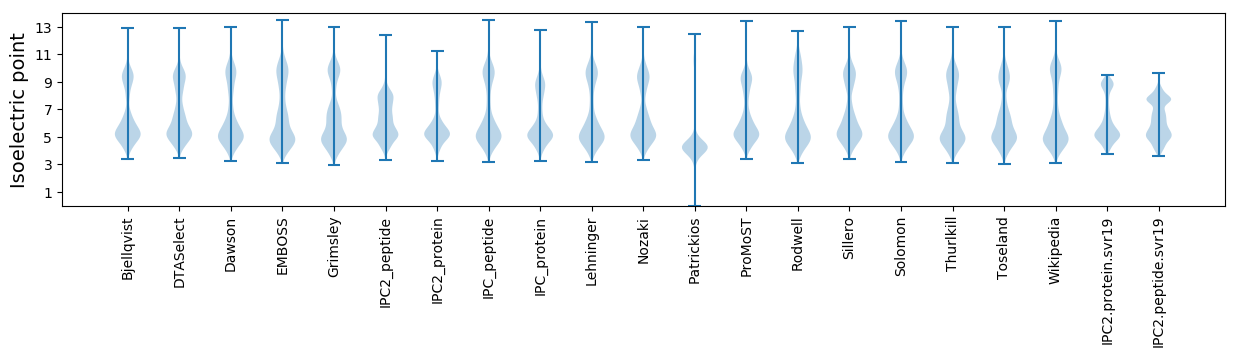
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A094WK42|A0A094WK42_ALKAL Cold-shock protein OS=Bacillus alcalophilus ATCC 27647 = CGMCC 1.3604 OX=1218173 GN=AJ85_03455 PE=4 SV=1
MM1 pKa = 7.71GKK3 pKa = 8.0PTFQPNNRR11 pKa = 11.84KK12 pKa = 9.23RR13 pKa = 11.84KK14 pKa = 8.22KK15 pKa = 8.69VHH17 pKa = 5.46GFRR20 pKa = 11.84ARR22 pKa = 11.84MSTKK26 pKa = 10.19NGRR29 pKa = 11.84KK30 pKa = 8.49VLARR34 pKa = 11.84RR35 pKa = 11.84RR36 pKa = 11.84RR37 pKa = 11.84KK38 pKa = 9.05GRR40 pKa = 11.84KK41 pKa = 8.7VLSAA45 pKa = 4.05
MM1 pKa = 7.71GKK3 pKa = 8.0PTFQPNNRR11 pKa = 11.84KK12 pKa = 9.23RR13 pKa = 11.84KK14 pKa = 8.22KK15 pKa = 8.69VHH17 pKa = 5.46GFRR20 pKa = 11.84ARR22 pKa = 11.84MSTKK26 pKa = 10.19NGRR29 pKa = 11.84KK30 pKa = 8.49VLARR34 pKa = 11.84RR35 pKa = 11.84RR36 pKa = 11.84RR37 pKa = 11.84KK38 pKa = 9.05GRR40 pKa = 11.84KK41 pKa = 8.7VLSAA45 pKa = 4.05
Molecular weight: 5.31 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1064280 |
26 |
1694 |
284.3 |
32.05 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.776 ± 0.045 | 0.69 ± 0.013 |
4.979 ± 0.031 | 8.167 ± 0.05 |
4.62 ± 0.03 | 6.656 ± 0.039 |
2.117 ± 0.019 | 7.722 ± 0.039 |
6.681 ± 0.04 | 10.094 ± 0.047 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.72 ± 0.019 | 4.325 ± 0.028 |
3.474 ± 0.021 | 4.054 ± 0.029 |
4.087 ± 0.031 | 5.922 ± 0.03 |
5.318 ± 0.024 | 7.068 ± 0.034 |
1.049 ± 0.015 | 3.481 ± 0.024 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
