
Beihai tombus-like virus 16
Taxonomy: Viruses; Riboviria; unclassified Riboviria; unclassified RNA viruses ShiM-2016
Average proteome isoelectric point is 7.66
Get precalculated fractions of proteins
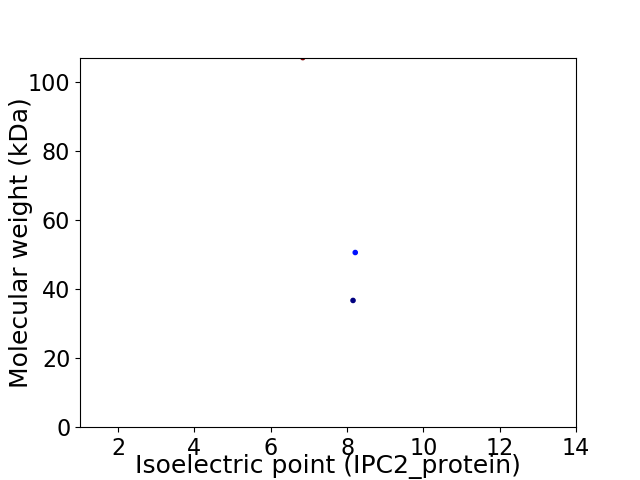
Virtual 2D-PAGE plot for 3 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions

Protein with the lowest isoelectric point:
>tr|A0A1L3KFS6|A0A1L3KFS6_9VIRU Uncharacterized protein OS=Beihai tombus-like virus 16 OX=1922719 PE=4 SV=1
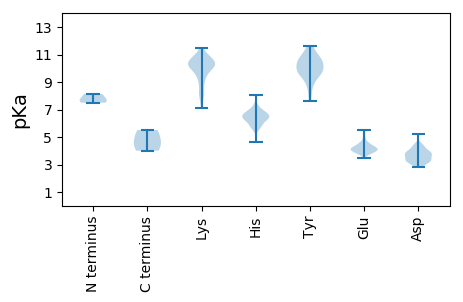
MM1 pKa = 8.13RR2 pKa = 11.84STKK5 pKa = 10.32INISFKK11 pKa = 10.33GPRR14 pKa = 11.84MDD16 pKa = 3.95YY17 pKa = 10.97LYY19 pKa = 10.93LKK21 pKa = 10.2KK22 pKa = 10.82LLGSNVNIQSTGNWEE37 pKa = 4.29EE38 pKa = 3.67MDD40 pKa = 3.73HH41 pKa = 6.46ARR43 pKa = 11.84LRR45 pKa = 11.84AFRR48 pKa = 11.84DD49 pKa = 3.47CCYY52 pKa = 10.6QWLLQQCRR60 pKa = 11.84DD61 pKa = 3.42KK62 pKa = 10.97RR63 pKa = 11.84ILEE66 pKa = 4.0IGPRR70 pKa = 11.84RR71 pKa = 11.84HH72 pKa = 6.35YY73 pKa = 10.51IPHH76 pKa = 6.3FQQAAIKK83 pKa = 9.43TNGKK87 pKa = 7.33PCYY90 pKa = 8.19YY91 pKa = 10.19TVDD94 pKa = 3.51KK95 pKa = 10.87TEE97 pKa = 4.04KK98 pKa = 10.63ADD100 pKa = 3.7FQGRR104 pKa = 11.84FQDD107 pKa = 3.82LRR109 pKa = 11.84HH110 pKa = 6.79DD111 pKa = 3.73NFDD114 pKa = 3.83IIFGVDD120 pKa = 3.24VYY122 pKa = 11.37DD123 pKa = 4.31LQDD126 pKa = 4.14FVPQNDD132 pKa = 2.9NVTYY136 pKa = 9.76YY137 pKa = 10.5FALAEE142 pKa = 4.27YY143 pKa = 9.38PHH145 pKa = 6.37TACGGTGKK153 pKa = 10.82LEE155 pKa = 4.18LPDD158 pKa = 4.14LGVIVTPDD166 pKa = 2.84GHH168 pKa = 5.95TTDD171 pKa = 5.19SGDD174 pKa = 3.41KK175 pKa = 10.61NYY177 pKa = 10.04TDD179 pKa = 4.36KK180 pKa = 11.47VPDD183 pKa = 3.93YY184 pKa = 9.98LTLQLPSFAKK194 pKa = 9.74EE195 pKa = 3.7HH196 pKa = 6.39WPSGWNIEE204 pKa = 4.2LVITYY209 pKa = 9.54GCYY212 pKa = 10.26SIYY215 pKa = 10.19KK216 pKa = 7.93VHH218 pKa = 6.34TGEE221 pKa = 4.52RR222 pKa = 11.84SPVSLKK228 pKa = 11.15SNTFFDD234 pKa = 4.56ASSSSYY240 pKa = 11.22GYY242 pKa = 9.55VCEE245 pKa = 4.07PTTVGAHH252 pKa = 6.32ICPAQLVNLAMPHH265 pKa = 5.56GRR267 pKa = 11.84DD268 pKa = 3.52YY269 pKa = 11.65NKK271 pKa = 10.27NAIRR275 pKa = 11.84AGTNALVDD283 pKa = 3.99EE284 pKa = 4.71ARR286 pKa = 11.84KK287 pKa = 9.99ISDD290 pKa = 4.01DD291 pKa = 3.64LFLTIEE297 pKa = 4.2QIRR300 pKa = 11.84HH301 pKa = 4.61YY302 pKa = 9.98ACSAVRR308 pKa = 11.84ISSLRR313 pKa = 11.84NAAEE317 pKa = 4.24HH318 pKa = 6.51ARR320 pKa = 11.84SANDD324 pKa = 2.95IYY326 pKa = 11.05TALAHH331 pKa = 6.17NARR334 pKa = 11.84GVTVQQNASLVARR347 pKa = 11.84LVDD350 pKa = 3.72QIEE353 pKa = 4.19FALEE357 pKa = 3.84FMNGDD362 pKa = 4.0AEE364 pKa = 4.24QHH366 pKa = 6.55PLTKK370 pKa = 10.58LLTKK374 pKa = 10.23ILTLLISTCKK384 pKa = 10.56NIARR388 pKa = 11.84ALRR391 pKa = 11.84DD392 pKa = 3.86LIEE395 pKa = 4.47TLGDD399 pKa = 4.28WIRR402 pKa = 11.84EE403 pKa = 3.95ACIWGLRR410 pKa = 11.84QNSISLIHH418 pKa = 6.88PLLKK422 pKa = 10.21WILDD426 pKa = 3.72QIHH429 pKa = 6.43PVSYY433 pKa = 10.53HH434 pKa = 5.72EE435 pKa = 4.44RR436 pKa = 11.84DD437 pKa = 3.4SVRR440 pKa = 11.84SHH442 pKa = 6.84LSSVPIAPDD451 pKa = 3.14EE452 pKa = 4.05TSNIHH457 pKa = 6.27NYY459 pKa = 10.26RR460 pKa = 11.84NIHH463 pKa = 5.55TEE465 pKa = 3.79ADD467 pKa = 3.99FKK469 pKa = 11.24PPSSYY474 pKa = 9.65TQLGPHH480 pKa = 5.93LVEE483 pKa = 4.54EE484 pKa = 4.53RR485 pKa = 11.84PHH487 pKa = 8.03VIVHH491 pKa = 6.87DD492 pKa = 3.86KK493 pKa = 10.56DD494 pKa = 3.68ARR496 pKa = 11.84DD497 pKa = 3.12AVIIQRR503 pKa = 11.84EE504 pKa = 4.31GRR506 pKa = 11.84PTPQGLDD513 pKa = 3.29YY514 pKa = 11.2SILEE518 pKa = 4.13EE519 pKa = 3.79TAEE522 pKa = 3.92QVGIEE527 pKa = 4.15LQSFITQEE535 pKa = 4.01TGQSSVEE542 pKa = 3.83PVDD545 pKa = 4.04FQFGSTVSQARR556 pKa = 11.84VAPSSMQPGEE566 pKa = 4.19TSTTEE571 pKa = 3.89QPATSPKK578 pKa = 8.91LTSSSSSSLSNLASSRR594 pKa = 11.84TSFLPGTFGHH604 pKa = 6.45VPPQVGTCLPSKK616 pKa = 9.33TLSKK620 pKa = 9.68TIQTLSVEE628 pKa = 4.49KK629 pKa = 10.16PSKK632 pKa = 10.26KK633 pKa = 8.97QLKK636 pKa = 9.57PFTKK640 pKa = 9.34IAKK643 pKa = 7.65PHH645 pKa = 6.36KK646 pKa = 9.69LGKK649 pKa = 8.21STNVTSPPSTPANPPKK665 pKa = 10.25YY666 pKa = 8.89YY667 pKa = 10.27KK668 pKa = 10.63SSTTSVTEE676 pKa = 4.0SCSDD680 pKa = 3.57MLPSCIKK687 pKa = 10.71LLTHH691 pKa = 5.92SCKK694 pKa = 10.57ASAPSPTTSEE704 pKa = 4.62DD705 pKa = 3.14GSLPSQRR712 pKa = 11.84EE713 pKa = 3.76GADD716 pKa = 3.35AQEE719 pKa = 4.54TPTPTSATPSSTTYY733 pKa = 8.95YY734 pKa = 9.38TAASRR739 pKa = 11.84KK740 pKa = 9.05LNPWSGGLAVMIALSSIITLMSPTLPVSALPPKK773 pKa = 10.07SAAAAVSGTPLSVLPWFIPQQEE795 pKa = 4.36GLHH798 pKa = 6.54FYY800 pKa = 9.38ATLINLLLGIVILLTIFLPKK820 pKa = 10.37LPEE823 pKa = 3.89FATKK827 pKa = 10.34KK828 pKa = 10.41NSQQKK833 pKa = 9.67PMPTMAYY840 pKa = 9.53QSSDD844 pKa = 3.2VCSIHH849 pKa = 6.98SPQNMALGFHH859 pKa = 6.67SLQTEE864 pKa = 4.06NPGICSYY871 pKa = 11.63DD872 pKa = 3.28AMEE875 pKa = 5.26NLPTYY880 pKa = 9.8NPPKK884 pKa = 9.83MLVSFLKK891 pKa = 10.94NILASHH897 pKa = 6.56SICNKK902 pKa = 8.85KK903 pKa = 10.63LKK905 pKa = 10.26IKK907 pKa = 10.73LDD909 pKa = 3.58DD910 pKa = 4.07HH911 pKa = 7.3QEE913 pKa = 3.89LLLICHH919 pKa = 7.34LSGSTSSPCRR929 pKa = 11.84PSIFRR934 pKa = 11.84GLLDD938 pKa = 4.45NIRR941 pKa = 11.84EE942 pKa = 4.11FPMALLDD949 pKa = 3.81KK950 pKa = 10.85LFNALIKK957 pKa = 10.87YY958 pKa = 9.92KK959 pKa = 10.75LEE961 pKa = 4.03SLLFSGG967 pKa = 5.51
MM1 pKa = 8.13RR2 pKa = 11.84STKK5 pKa = 10.32INISFKK11 pKa = 10.33GPRR14 pKa = 11.84MDD16 pKa = 3.95YY17 pKa = 10.97LYY19 pKa = 10.93LKK21 pKa = 10.2KK22 pKa = 10.82LLGSNVNIQSTGNWEE37 pKa = 4.29EE38 pKa = 3.67MDD40 pKa = 3.73HH41 pKa = 6.46ARR43 pKa = 11.84LRR45 pKa = 11.84AFRR48 pKa = 11.84DD49 pKa = 3.47CCYY52 pKa = 10.6QWLLQQCRR60 pKa = 11.84DD61 pKa = 3.42KK62 pKa = 10.97RR63 pKa = 11.84ILEE66 pKa = 4.0IGPRR70 pKa = 11.84RR71 pKa = 11.84HH72 pKa = 6.35YY73 pKa = 10.51IPHH76 pKa = 6.3FQQAAIKK83 pKa = 9.43TNGKK87 pKa = 7.33PCYY90 pKa = 8.19YY91 pKa = 10.19TVDD94 pKa = 3.51KK95 pKa = 10.87TEE97 pKa = 4.04KK98 pKa = 10.63ADD100 pKa = 3.7FQGRR104 pKa = 11.84FQDD107 pKa = 3.82LRR109 pKa = 11.84HH110 pKa = 6.79DD111 pKa = 3.73NFDD114 pKa = 3.83IIFGVDD120 pKa = 3.24VYY122 pKa = 11.37DD123 pKa = 4.31LQDD126 pKa = 4.14FVPQNDD132 pKa = 2.9NVTYY136 pKa = 9.76YY137 pKa = 10.5FALAEE142 pKa = 4.27YY143 pKa = 9.38PHH145 pKa = 6.37TACGGTGKK153 pKa = 10.82LEE155 pKa = 4.18LPDD158 pKa = 4.14LGVIVTPDD166 pKa = 2.84GHH168 pKa = 5.95TTDD171 pKa = 5.19SGDD174 pKa = 3.41KK175 pKa = 10.61NYY177 pKa = 10.04TDD179 pKa = 4.36KK180 pKa = 11.47VPDD183 pKa = 3.93YY184 pKa = 9.98LTLQLPSFAKK194 pKa = 9.74EE195 pKa = 3.7HH196 pKa = 6.39WPSGWNIEE204 pKa = 4.2LVITYY209 pKa = 9.54GCYY212 pKa = 10.26SIYY215 pKa = 10.19KK216 pKa = 7.93VHH218 pKa = 6.34TGEE221 pKa = 4.52RR222 pKa = 11.84SPVSLKK228 pKa = 11.15SNTFFDD234 pKa = 4.56ASSSSYY240 pKa = 11.22GYY242 pKa = 9.55VCEE245 pKa = 4.07PTTVGAHH252 pKa = 6.32ICPAQLVNLAMPHH265 pKa = 5.56GRR267 pKa = 11.84DD268 pKa = 3.52YY269 pKa = 11.65NKK271 pKa = 10.27NAIRR275 pKa = 11.84AGTNALVDD283 pKa = 3.99EE284 pKa = 4.71ARR286 pKa = 11.84KK287 pKa = 9.99ISDD290 pKa = 4.01DD291 pKa = 3.64LFLTIEE297 pKa = 4.2QIRR300 pKa = 11.84HH301 pKa = 4.61YY302 pKa = 9.98ACSAVRR308 pKa = 11.84ISSLRR313 pKa = 11.84NAAEE317 pKa = 4.24HH318 pKa = 6.51ARR320 pKa = 11.84SANDD324 pKa = 2.95IYY326 pKa = 11.05TALAHH331 pKa = 6.17NARR334 pKa = 11.84GVTVQQNASLVARR347 pKa = 11.84LVDD350 pKa = 3.72QIEE353 pKa = 4.19FALEE357 pKa = 3.84FMNGDD362 pKa = 4.0AEE364 pKa = 4.24QHH366 pKa = 6.55PLTKK370 pKa = 10.58LLTKK374 pKa = 10.23ILTLLISTCKK384 pKa = 10.56NIARR388 pKa = 11.84ALRR391 pKa = 11.84DD392 pKa = 3.86LIEE395 pKa = 4.47TLGDD399 pKa = 4.28WIRR402 pKa = 11.84EE403 pKa = 3.95ACIWGLRR410 pKa = 11.84QNSISLIHH418 pKa = 6.88PLLKK422 pKa = 10.21WILDD426 pKa = 3.72QIHH429 pKa = 6.43PVSYY433 pKa = 10.53HH434 pKa = 5.72EE435 pKa = 4.44RR436 pKa = 11.84DD437 pKa = 3.4SVRR440 pKa = 11.84SHH442 pKa = 6.84LSSVPIAPDD451 pKa = 3.14EE452 pKa = 4.05TSNIHH457 pKa = 6.27NYY459 pKa = 10.26RR460 pKa = 11.84NIHH463 pKa = 5.55TEE465 pKa = 3.79ADD467 pKa = 3.99FKK469 pKa = 11.24PPSSYY474 pKa = 9.65TQLGPHH480 pKa = 5.93LVEE483 pKa = 4.54EE484 pKa = 4.53RR485 pKa = 11.84PHH487 pKa = 8.03VIVHH491 pKa = 6.87DD492 pKa = 3.86KK493 pKa = 10.56DD494 pKa = 3.68ARR496 pKa = 11.84DD497 pKa = 3.12AVIIQRR503 pKa = 11.84EE504 pKa = 4.31GRR506 pKa = 11.84PTPQGLDD513 pKa = 3.29YY514 pKa = 11.2SILEE518 pKa = 4.13EE519 pKa = 3.79TAEE522 pKa = 3.92QVGIEE527 pKa = 4.15LQSFITQEE535 pKa = 4.01TGQSSVEE542 pKa = 3.83PVDD545 pKa = 4.04FQFGSTVSQARR556 pKa = 11.84VAPSSMQPGEE566 pKa = 4.19TSTTEE571 pKa = 3.89QPATSPKK578 pKa = 8.91LTSSSSSSLSNLASSRR594 pKa = 11.84TSFLPGTFGHH604 pKa = 6.45VPPQVGTCLPSKK616 pKa = 9.33TLSKK620 pKa = 9.68TIQTLSVEE628 pKa = 4.49KK629 pKa = 10.16PSKK632 pKa = 10.26KK633 pKa = 8.97QLKK636 pKa = 9.57PFTKK640 pKa = 9.34IAKK643 pKa = 7.65PHH645 pKa = 6.36KK646 pKa = 9.69LGKK649 pKa = 8.21STNVTSPPSTPANPPKK665 pKa = 10.25YY666 pKa = 8.89YY667 pKa = 10.27KK668 pKa = 10.63SSTTSVTEE676 pKa = 4.0SCSDD680 pKa = 3.57MLPSCIKK687 pKa = 10.71LLTHH691 pKa = 5.92SCKK694 pKa = 10.57ASAPSPTTSEE704 pKa = 4.62DD705 pKa = 3.14GSLPSQRR712 pKa = 11.84EE713 pKa = 3.76GADD716 pKa = 3.35AQEE719 pKa = 4.54TPTPTSATPSSTTYY733 pKa = 8.95YY734 pKa = 9.38TAASRR739 pKa = 11.84KK740 pKa = 9.05LNPWSGGLAVMIALSSIITLMSPTLPVSALPPKK773 pKa = 10.07SAAAAVSGTPLSVLPWFIPQQEE795 pKa = 4.36GLHH798 pKa = 6.54FYY800 pKa = 9.38ATLINLLLGIVILLTIFLPKK820 pKa = 10.37LPEE823 pKa = 3.89FATKK827 pKa = 10.34KK828 pKa = 10.41NSQQKK833 pKa = 9.67PMPTMAYY840 pKa = 9.53QSSDD844 pKa = 3.2VCSIHH849 pKa = 6.98SPQNMALGFHH859 pKa = 6.67SLQTEE864 pKa = 4.06NPGICSYY871 pKa = 11.63DD872 pKa = 3.28AMEE875 pKa = 5.26NLPTYY880 pKa = 9.8NPPKK884 pKa = 9.83MLVSFLKK891 pKa = 10.94NILASHH897 pKa = 6.56SICNKK902 pKa = 8.85KK903 pKa = 10.63LKK905 pKa = 10.26IKK907 pKa = 10.73LDD909 pKa = 3.58DD910 pKa = 4.07HH911 pKa = 7.3QEE913 pKa = 3.89LLLICHH919 pKa = 7.34LSGSTSSPCRR929 pKa = 11.84PSIFRR934 pKa = 11.84GLLDD938 pKa = 4.45NIRR941 pKa = 11.84EE942 pKa = 4.11FPMALLDD949 pKa = 3.81KK950 pKa = 10.85LFNALIKK957 pKa = 10.87YY958 pKa = 9.92KK959 pKa = 10.75LEE961 pKa = 4.03SLLFSGG967 pKa = 5.51
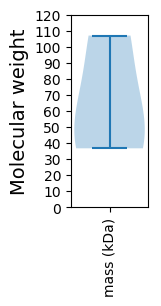
Molecular weight: 106.97 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1L3KFS6|A0A1L3KFS6_9VIRU Uncharacterized protein OS=Beihai tombus-like virus 16 OX=1922719 PE=4 SV=1
MM1 pKa = 7.5TLQFHH6 pKa = 7.12LLLKK10 pKa = 9.95IHH12 pKa = 6.52NFLICNMKK20 pKa = 10.23LKK22 pKa = 10.37KK23 pKa = 10.02KK24 pKa = 8.63QQQRR28 pKa = 11.84PKK30 pKa = 10.16KK31 pKa = 9.29APKK34 pKa = 10.06KK35 pKa = 8.94KK36 pKa = 7.11TTSKK40 pKa = 10.6PSRR43 pKa = 11.84QLNDD47 pKa = 3.39GLKK50 pKa = 10.15VRR52 pKa = 11.84YY53 pKa = 8.93KK54 pKa = 9.31EE55 pKa = 3.88CWGKK59 pKa = 10.33VEE61 pKa = 5.53SGFNSYY67 pKa = 10.6KK68 pKa = 10.38FLPGSSGLKK77 pKa = 10.38HH78 pKa = 6.83LDD80 pKa = 3.17SLAAIYY86 pKa = 9.77EE87 pKa = 4.46SYY89 pKa = 10.71KK90 pKa = 10.12ILSTVRR96 pKa = 11.84VSFASSTSATTSGEE110 pKa = 3.46IVMGVDD116 pKa = 3.04YY117 pKa = 10.67DD118 pKa = 3.89ARR120 pKa = 11.84DD121 pKa = 4.01EE122 pKa = 4.21ISDD125 pKa = 3.56FHH127 pKa = 6.74GVCALQPKK135 pKa = 9.67HH136 pKa = 6.25VGPVRR141 pKa = 11.84NAGNLSVPHH150 pKa = 5.79TRR152 pKa = 11.84AMHH155 pKa = 5.74KK156 pKa = 10.05KK157 pKa = 8.25WLFTNSSAAPGEE169 pKa = 4.04AAFAIHH175 pKa = 6.78VYY177 pKa = 10.6NGAAEE182 pKa = 4.35SAGTMWVEE190 pKa = 3.79YY191 pKa = 9.4DD192 pKa = 3.52LHH194 pKa = 6.8FISPTTKK201 pKa = 10.13ASLVSVFEE209 pKa = 4.23SFEE212 pKa = 4.21VPASSGGGVANRR224 pKa = 11.84PIYY227 pKa = 10.56SSIRR231 pKa = 11.84MLGNTAYY238 pKa = 10.57NEE240 pKa = 4.1KK241 pKa = 10.18IAANTVLWVVSEE253 pKa = 4.38YY254 pKa = 11.04DD255 pKa = 3.55VQINTIFWTILEE267 pKa = 4.17SLTRR271 pKa = 11.84YY272 pKa = 9.88IEE274 pKa = 3.99GRR276 pKa = 11.84PYY278 pKa = 10.21KK279 pKa = 10.47VKK281 pKa = 10.31AVKK284 pKa = 10.11AIKK287 pKa = 8.3EE288 pKa = 4.54TQDD291 pKa = 3.26VPATCSGSGNFWAIATSAYY310 pKa = 10.22AAYY313 pKa = 7.81TALIACWSVHH323 pKa = 5.91FDD325 pKa = 4.37LPKK328 pKa = 10.29KK329 pKa = 10.07DD330 pKa = 4.36APAPTPIVSPTTLKK344 pKa = 10.9EE345 pKa = 4.04PFSNPSPTRR354 pKa = 11.84EE355 pKa = 4.93DD356 pKa = 2.89INAQAKK362 pKa = 8.24LQSYY366 pKa = 7.64PKK368 pKa = 9.84KK369 pKa = 9.72KK370 pKa = 10.19RR371 pKa = 11.84EE372 pKa = 3.83MTRR375 pKa = 11.84AEE377 pKa = 3.88AEE379 pKa = 3.74EE380 pKa = 4.74AIRR383 pKa = 11.84KK384 pKa = 7.74SVRR387 pKa = 11.84DD388 pKa = 3.16ADD390 pKa = 3.68AARR393 pKa = 11.84WNSIVEE399 pKa = 4.12FFSRR403 pKa = 11.84GNLQRR408 pKa = 11.84TLQQDD413 pKa = 3.1LDD415 pKa = 3.89KK416 pKa = 11.39AQNCPTTLIRR426 pKa = 11.84DD427 pKa = 3.78DD428 pKa = 3.89PTPVLEE434 pKa = 4.25KK435 pKa = 10.85QEE437 pKa = 4.39LEE439 pKa = 4.21SPLEE443 pKa = 4.01FTEE446 pKa = 4.97TSCQLTIDD454 pKa = 3.87EE455 pKa = 4.67
MM1 pKa = 7.5TLQFHH6 pKa = 7.12LLLKK10 pKa = 9.95IHH12 pKa = 6.52NFLICNMKK20 pKa = 10.23LKK22 pKa = 10.37KK23 pKa = 10.02KK24 pKa = 8.63QQQRR28 pKa = 11.84PKK30 pKa = 10.16KK31 pKa = 9.29APKK34 pKa = 10.06KK35 pKa = 8.94KK36 pKa = 7.11TTSKK40 pKa = 10.6PSRR43 pKa = 11.84QLNDD47 pKa = 3.39GLKK50 pKa = 10.15VRR52 pKa = 11.84YY53 pKa = 8.93KK54 pKa = 9.31EE55 pKa = 3.88CWGKK59 pKa = 10.33VEE61 pKa = 5.53SGFNSYY67 pKa = 10.6KK68 pKa = 10.38FLPGSSGLKK77 pKa = 10.38HH78 pKa = 6.83LDD80 pKa = 3.17SLAAIYY86 pKa = 9.77EE87 pKa = 4.46SYY89 pKa = 10.71KK90 pKa = 10.12ILSTVRR96 pKa = 11.84VSFASSTSATTSGEE110 pKa = 3.46IVMGVDD116 pKa = 3.04YY117 pKa = 10.67DD118 pKa = 3.89ARR120 pKa = 11.84DD121 pKa = 4.01EE122 pKa = 4.21ISDD125 pKa = 3.56FHH127 pKa = 6.74GVCALQPKK135 pKa = 9.67HH136 pKa = 6.25VGPVRR141 pKa = 11.84NAGNLSVPHH150 pKa = 5.79TRR152 pKa = 11.84AMHH155 pKa = 5.74KK156 pKa = 10.05KK157 pKa = 8.25WLFTNSSAAPGEE169 pKa = 4.04AAFAIHH175 pKa = 6.78VYY177 pKa = 10.6NGAAEE182 pKa = 4.35SAGTMWVEE190 pKa = 3.79YY191 pKa = 9.4DD192 pKa = 3.52LHH194 pKa = 6.8FISPTTKK201 pKa = 10.13ASLVSVFEE209 pKa = 4.23SFEE212 pKa = 4.21VPASSGGGVANRR224 pKa = 11.84PIYY227 pKa = 10.56SSIRR231 pKa = 11.84MLGNTAYY238 pKa = 10.57NEE240 pKa = 4.1KK241 pKa = 10.18IAANTVLWVVSEE253 pKa = 4.38YY254 pKa = 11.04DD255 pKa = 3.55VQINTIFWTILEE267 pKa = 4.17SLTRR271 pKa = 11.84YY272 pKa = 9.88IEE274 pKa = 3.99GRR276 pKa = 11.84PYY278 pKa = 10.21KK279 pKa = 10.47VKK281 pKa = 10.31AVKK284 pKa = 10.11AIKK287 pKa = 8.3EE288 pKa = 4.54TQDD291 pKa = 3.26VPATCSGSGNFWAIATSAYY310 pKa = 10.22AAYY313 pKa = 7.81TALIACWSVHH323 pKa = 5.91FDD325 pKa = 4.37LPKK328 pKa = 10.29KK329 pKa = 10.07DD330 pKa = 4.36APAPTPIVSPTTLKK344 pKa = 10.9EE345 pKa = 4.04PFSNPSPTRR354 pKa = 11.84EE355 pKa = 4.93DD356 pKa = 2.89INAQAKK362 pKa = 8.24LQSYY366 pKa = 7.64PKK368 pKa = 9.84KK369 pKa = 9.72KK370 pKa = 10.19RR371 pKa = 11.84EE372 pKa = 3.83MTRR375 pKa = 11.84AEE377 pKa = 3.88AEE379 pKa = 3.74EE380 pKa = 4.74AIRR383 pKa = 11.84KK384 pKa = 7.74SVRR387 pKa = 11.84DD388 pKa = 3.16ADD390 pKa = 3.68AARR393 pKa = 11.84WNSIVEE399 pKa = 4.12FFSRR403 pKa = 11.84GNLQRR408 pKa = 11.84TLQQDD413 pKa = 3.1LDD415 pKa = 3.89KK416 pKa = 11.39AQNCPTTLIRR426 pKa = 11.84DD427 pKa = 3.78DD428 pKa = 3.89PTPVLEE434 pKa = 4.25KK435 pKa = 10.85QEE437 pKa = 4.39LEE439 pKa = 4.21SPLEE443 pKa = 4.01FTEE446 pKa = 4.97TSCQLTIDD454 pKa = 3.87EE455 pKa = 4.67
Molecular weight: 50.56 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1744 |
322 |
967 |
581.3 |
64.74 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.683 ± 0.765 | 2.122 ± 0.272 |
4.759 ± 0.217 | 4.931 ± 0.446 |
4.014 ± 0.577 | 4.874 ± 0.116 |
2.81 ± 0.346 | 5.963 ± 0.254 |
6.135 ± 0.66 | 9.346 ± 0.783 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.433 ± 0.123 | 4.186 ± 0.124 |
6.25 ± 0.701 | 4.186 ± 0.251 |
4.644 ± 0.551 | 9.576 ± 0.551 |
7.454 ± 0.08 | 4.817 ± 0.513 |
1.089 ± 0.244 | 3.727 ± 0.38 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
