
Tortoise microvirus 50
Taxonomy: Viruses; Monodnaviria; Sangervirae; Phixviricota; Malgrandaviricetes; Petitvirales; Microviridae; unclassified Microviridae
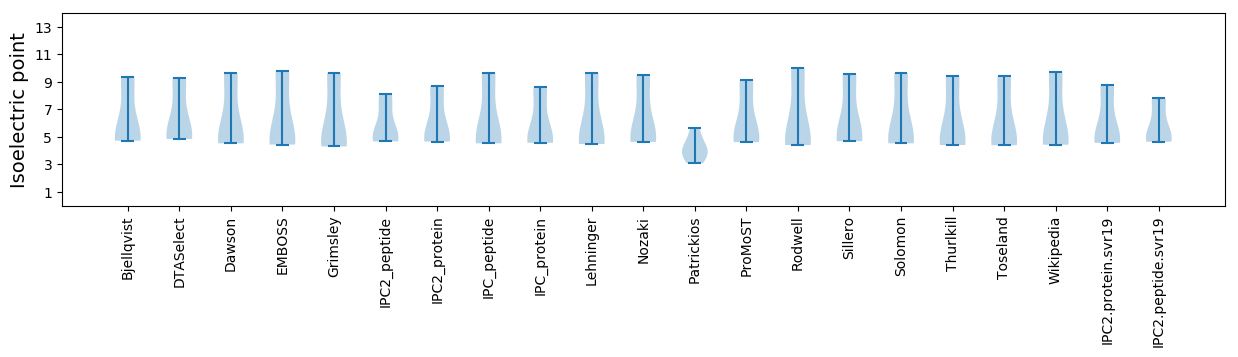
Average proteome isoelectric point is 6.11
Get precalculated fractions of proteins
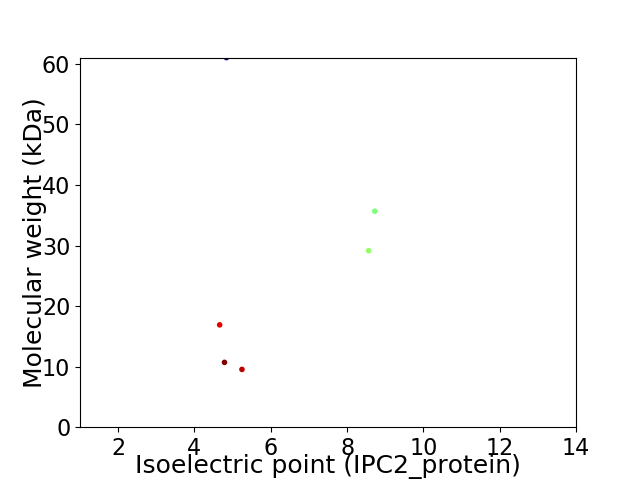
Virtual 2D-PAGE plot for 6 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A4P8W7D3|A0A4P8W7D3_9VIRU DNA pilot protein OS=Tortoise microvirus 50 OX=2583155 PE=4 SV=1
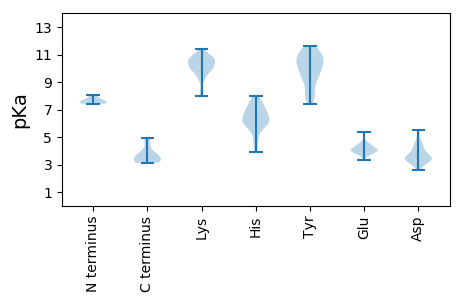
MM1 pKa = 7.74FDD3 pKa = 3.52FSYY6 pKa = 11.25SVFLLSDD13 pKa = 3.56SVRR16 pKa = 11.84YY17 pKa = 10.01DD18 pKa = 3.11SFPVLKK24 pKa = 9.88EE25 pKa = 3.67YY26 pKa = 11.14AKK28 pKa = 9.27THH30 pKa = 5.2QPFIGFNDD38 pKa = 3.33YY39 pKa = 11.14RR40 pKa = 11.84HH41 pKa = 6.29FLTFLEE47 pKa = 4.61TTDD50 pKa = 3.63CQKK53 pKa = 11.07FILLVSKK60 pKa = 10.29FEE62 pKa = 4.28NDD64 pKa = 3.02EE65 pKa = 4.14NPAFSSFIEE74 pKa = 4.12VFDD77 pKa = 4.58PVSCTCVKK85 pKa = 10.63SFEE88 pKa = 4.25HH89 pKa = 7.07SII91 pKa = 3.56
MM1 pKa = 7.74FDD3 pKa = 3.52FSYY6 pKa = 11.25SVFLLSDD13 pKa = 3.56SVRR16 pKa = 11.84YY17 pKa = 10.01DD18 pKa = 3.11SFPVLKK24 pKa = 9.88EE25 pKa = 3.67YY26 pKa = 11.14AKK28 pKa = 9.27THH30 pKa = 5.2QPFIGFNDD38 pKa = 3.33YY39 pKa = 11.14RR40 pKa = 11.84HH41 pKa = 6.29FLTFLEE47 pKa = 4.61TTDD50 pKa = 3.63CQKK53 pKa = 11.07FILLVSKK60 pKa = 10.29FEE62 pKa = 4.28NDD64 pKa = 3.02EE65 pKa = 4.14NPAFSSFIEE74 pKa = 4.12VFDD77 pKa = 4.58PVSCTCVKK85 pKa = 10.63SFEE88 pKa = 4.25HH89 pKa = 7.07SII91 pKa = 3.56
Molecular weight: 10.71 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A4P8W9V4|A0A4P8W9V4_9VIRU Replication initiation protein OS=Tortoise microvirus 50 OX=2583155 PE=4 SV=1
MM1 pKa = 7.61CNSPLVAMKK10 pKa = 10.77LEE12 pKa = 4.0NVYY15 pKa = 9.55TKK17 pKa = 10.38SGKK20 pKa = 10.05NKK22 pKa = 9.29VVVLGGLAARR32 pKa = 11.84EE33 pKa = 3.93HH34 pKa = 7.17LEE36 pKa = 4.51FGDD39 pKa = 4.09SSQLNTPFLIPCGKK53 pKa = 9.97CVSCRR58 pKa = 11.84IKK60 pKa = 10.51HH61 pKa = 4.66SRR63 pKa = 11.84EE64 pKa = 3.37WAVRR68 pKa = 11.84CCHH71 pKa = 5.99EE72 pKa = 4.1ASLYY76 pKa = 10.3EE77 pKa = 4.19KK78 pKa = 10.84NCFITLTYY86 pKa = 10.81NNEE89 pKa = 4.13HH90 pKa = 6.47LPSDD94 pKa = 3.45KK95 pKa = 10.74SLHH98 pKa = 6.01LEE100 pKa = 3.77HH101 pKa = 6.58WQKK104 pKa = 10.74FMKK107 pKa = 10.37RR108 pKa = 11.84FRR110 pKa = 11.84KK111 pKa = 10.0KK112 pKa = 8.27YY113 pKa = 9.14GEE115 pKa = 4.32GIRR118 pKa = 11.84FFMCGEE124 pKa = 4.07YY125 pKa = 10.93GSVNARR131 pKa = 11.84PHH133 pKa = 4.48YY134 pKa = 9.11HH135 pKa = 6.11ACIFGFDD142 pKa = 4.08FPDD145 pKa = 4.09KK146 pKa = 10.54EE147 pKa = 3.75IWQVRR152 pKa = 11.84EE153 pKa = 3.99KK154 pKa = 10.6IPLYY158 pKa = 10.07RR159 pKa = 11.84SHH161 pKa = 7.93SLEE164 pKa = 4.0KK165 pKa = 10.09LWPFGFSTIGEE176 pKa = 4.45VTFEE180 pKa = 3.97SAAYY184 pKa = 7.43VARR187 pKa = 11.84YY188 pKa = 8.75IFKK191 pKa = 10.74KK192 pKa = 9.08ITGDD196 pKa = 3.41KK197 pKa = 10.61AAEE200 pKa = 4.31HH201 pKa = 5.94YY202 pKa = 10.51NGRR205 pKa = 11.84KK206 pKa = 9.4PEE208 pKa = 4.72FITMSRR214 pKa = 11.84RR215 pKa = 11.84PGIAHH220 pKa = 7.22DD221 pKa = 3.64WFLKK225 pKa = 9.88NEE227 pKa = 3.92RR228 pKa = 11.84DD229 pKa = 3.37IYY231 pKa = 11.62SNDD234 pKa = 3.1FLIDD238 pKa = 3.58QKK240 pKa = 10.81KK241 pKa = 10.02HH242 pKa = 6.01KK243 pKa = 9.81IRR245 pKa = 11.84PLRR248 pKa = 11.84YY249 pKa = 7.92YY250 pKa = 10.44DD251 pKa = 4.11HH252 pKa = 7.98LMEE255 pKa = 5.35IYY257 pKa = 10.68SPDD260 pKa = 3.08EE261 pKa = 3.72YY262 pKa = 11.51YY263 pKa = 10.09ILKK266 pKa = 9.69KK267 pKa = 10.33QRR269 pKa = 11.84LEE271 pKa = 3.93NLSKK275 pKa = 10.12IDD277 pKa = 3.31SFQRR281 pKa = 11.84MQDD284 pKa = 2.94KK285 pKa = 10.87EE286 pKa = 4.26KK287 pKa = 10.68FKK289 pKa = 10.78KK290 pKa = 9.98YY291 pKa = 8.07QLKK294 pKa = 10.69KK295 pKa = 10.44LVRR298 pKa = 11.84PLL300 pKa = 3.36
MM1 pKa = 7.61CNSPLVAMKK10 pKa = 10.77LEE12 pKa = 4.0NVYY15 pKa = 9.55TKK17 pKa = 10.38SGKK20 pKa = 10.05NKK22 pKa = 9.29VVVLGGLAARR32 pKa = 11.84EE33 pKa = 3.93HH34 pKa = 7.17LEE36 pKa = 4.51FGDD39 pKa = 4.09SSQLNTPFLIPCGKK53 pKa = 9.97CVSCRR58 pKa = 11.84IKK60 pKa = 10.51HH61 pKa = 4.66SRR63 pKa = 11.84EE64 pKa = 3.37WAVRR68 pKa = 11.84CCHH71 pKa = 5.99EE72 pKa = 4.1ASLYY76 pKa = 10.3EE77 pKa = 4.19KK78 pKa = 10.84NCFITLTYY86 pKa = 10.81NNEE89 pKa = 4.13HH90 pKa = 6.47LPSDD94 pKa = 3.45KK95 pKa = 10.74SLHH98 pKa = 6.01LEE100 pKa = 3.77HH101 pKa = 6.58WQKK104 pKa = 10.74FMKK107 pKa = 10.37RR108 pKa = 11.84FRR110 pKa = 11.84KK111 pKa = 10.0KK112 pKa = 8.27YY113 pKa = 9.14GEE115 pKa = 4.32GIRR118 pKa = 11.84FFMCGEE124 pKa = 4.07YY125 pKa = 10.93GSVNARR131 pKa = 11.84PHH133 pKa = 4.48YY134 pKa = 9.11HH135 pKa = 6.11ACIFGFDD142 pKa = 4.08FPDD145 pKa = 4.09KK146 pKa = 10.54EE147 pKa = 3.75IWQVRR152 pKa = 11.84EE153 pKa = 3.99KK154 pKa = 10.6IPLYY158 pKa = 10.07RR159 pKa = 11.84SHH161 pKa = 7.93SLEE164 pKa = 4.0KK165 pKa = 10.09LWPFGFSTIGEE176 pKa = 4.45VTFEE180 pKa = 3.97SAAYY184 pKa = 7.43VARR187 pKa = 11.84YY188 pKa = 8.75IFKK191 pKa = 10.74KK192 pKa = 9.08ITGDD196 pKa = 3.41KK197 pKa = 10.61AAEE200 pKa = 4.31HH201 pKa = 5.94YY202 pKa = 10.51NGRR205 pKa = 11.84KK206 pKa = 9.4PEE208 pKa = 4.72FITMSRR214 pKa = 11.84RR215 pKa = 11.84PGIAHH220 pKa = 7.22DD221 pKa = 3.64WFLKK225 pKa = 9.88NEE227 pKa = 3.92RR228 pKa = 11.84DD229 pKa = 3.37IYY231 pKa = 11.62SNDD234 pKa = 3.1FLIDD238 pKa = 3.58QKK240 pKa = 10.81KK241 pKa = 10.02HH242 pKa = 6.01KK243 pKa = 9.81IRR245 pKa = 11.84PLRR248 pKa = 11.84YY249 pKa = 7.92YY250 pKa = 10.44DD251 pKa = 4.11HH252 pKa = 7.98LMEE255 pKa = 5.35IYY257 pKa = 10.68SPDD260 pKa = 3.08EE261 pKa = 3.72YY262 pKa = 11.51YY263 pKa = 10.09ILKK266 pKa = 9.69KK267 pKa = 10.33QRR269 pKa = 11.84LEE271 pKa = 3.93NLSKK275 pKa = 10.12IDD277 pKa = 3.31SFQRR281 pKa = 11.84MQDD284 pKa = 2.94KK285 pKa = 10.87EE286 pKa = 4.26KK287 pKa = 10.68FKK289 pKa = 10.78KK290 pKa = 9.98YY291 pKa = 8.07QLKK294 pKa = 10.69KK295 pKa = 10.44LVRR298 pKa = 11.84PLL300 pKa = 3.36
Molecular weight: 35.67 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1443 |
85 |
548 |
240.5 |
27.16 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.9 ± 2.204 | 1.317 ± 0.708 |
6.029 ± 1.087 | 5.613 ± 1.167 |
6.376 ± 1.478 | 5.128 ± 1.045 |
2.287 ± 0.583 | 5.128 ± 0.261 |
5.752 ± 1.955 | 8.593 ± 0.337 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.455 ± 0.257 | 5.059 ± 0.546 |
4.782 ± 0.79 | 4.019 ± 0.606 |
5.128 ± 0.544 | 9.979 ± 1.526 |
4.782 ± 0.599 | 5.267 ± 0.52 |
1.386 ± 0.25 | 4.019 ± 0.63 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
