
Capybara microvirus Cap3_SP_423
Taxonomy: Viruses; Monodnaviria; Sangervirae; Phixviricota; Malgrandaviricetes; Petitvirales; Microviridae; unclassified Microviridae
Average proteome isoelectric point is 5.82
Get precalculated fractions of proteins
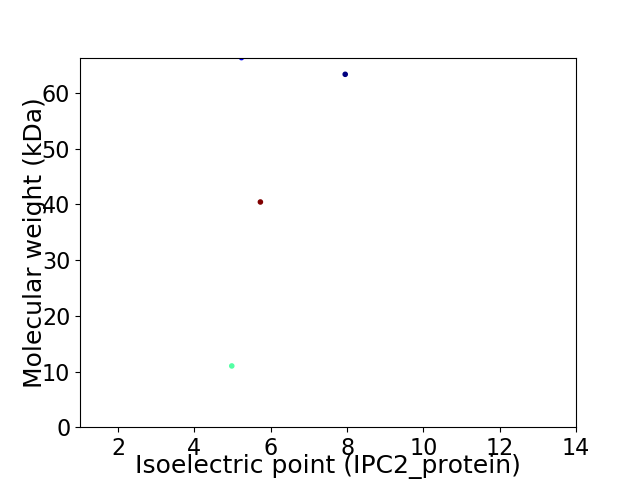
Virtual 2D-PAGE plot for 4 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A4P8W8J5|A0A4P8W8J5_9VIRU Minor capsid protein OS=Capybara microvirus Cap3_SP_423 OX=2585449 PE=4 SV=1
MM1 pKa = 7.6LFNLKK6 pKa = 10.21YY7 pKa = 10.6YY8 pKa = 10.02NKK10 pKa = 10.0CEE12 pKa = 4.09NPEE15 pKa = 4.12FPPLLVPQKK24 pKa = 9.51TIVEE28 pKa = 4.22GSEE31 pKa = 3.61EE32 pKa = 4.59VIFLEE37 pKa = 5.25HH38 pKa = 6.5VPQNDD43 pKa = 4.12FPSEE47 pKa = 4.12MYY49 pKa = 10.66DD50 pKa = 3.66LQNLKK55 pKa = 10.53SAGISPTQFRR65 pKa = 11.84GGSVPSTSRR74 pKa = 11.84LSDD77 pKa = 3.19QSKK80 pKa = 11.05ADD82 pKa = 4.22AIVKK86 pKa = 10.36NIVNALNTKK95 pKa = 9.1EE96 pKa = 4.13NEE98 pKa = 3.98
MM1 pKa = 7.6LFNLKK6 pKa = 10.21YY7 pKa = 10.6YY8 pKa = 10.02NKK10 pKa = 10.0CEE12 pKa = 4.09NPEE15 pKa = 4.12FPPLLVPQKK24 pKa = 9.51TIVEE28 pKa = 4.22GSEE31 pKa = 3.61EE32 pKa = 4.59VIFLEE37 pKa = 5.25HH38 pKa = 6.5VPQNDD43 pKa = 4.12FPSEE47 pKa = 4.12MYY49 pKa = 10.66DD50 pKa = 3.66LQNLKK55 pKa = 10.53SAGISPTQFRR65 pKa = 11.84GGSVPSTSRR74 pKa = 11.84LSDD77 pKa = 3.19QSKK80 pKa = 11.05ADD82 pKa = 4.22AIVKK86 pKa = 10.36NIVNALNTKK95 pKa = 9.1EE96 pKa = 4.13NEE98 pKa = 3.98
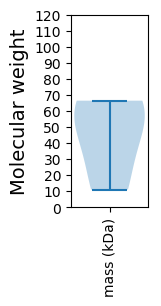
Molecular weight: 11.0 kDa
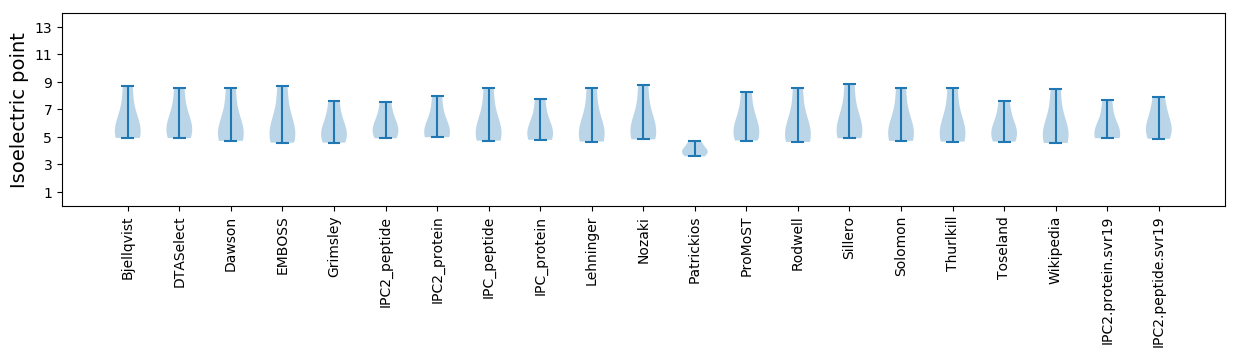
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A4P8W627|A0A4P8W627_9VIRU Uncharacterized protein OS=Capybara microvirus Cap3_SP_423 OX=2585449 PE=4 SV=1
MM1 pKa = 8.06RR2 pKa = 11.84LRR4 pKa = 11.84NRR6 pKa = 11.84ITIPTAFAYY15 pKa = 10.09QSQKK19 pKa = 10.29RR20 pKa = 11.84MEE22 pKa = 4.13YY23 pKa = 9.82TIRR26 pKa = 11.84AYY28 pKa = 10.65YY29 pKa = 9.75QFEE32 pKa = 4.08EE33 pKa = 4.59TKK35 pKa = 10.9LKK37 pKa = 10.77GGQQFFYY44 pKa = 10.55TLTYY48 pKa = 10.9NDD50 pKa = 3.69DD51 pKa = 3.92HH52 pKa = 7.63LPTFLGRR59 pKa = 11.84NCHH62 pKa = 6.96DD63 pKa = 4.04YY64 pKa = 11.75NDD66 pKa = 3.44IRR68 pKa = 11.84YY69 pKa = 7.86LTVGAFAKK77 pKa = 10.56KK78 pKa = 10.2LLRR81 pKa = 11.84HH82 pKa = 6.08YY83 pKa = 9.27NLSMKK88 pKa = 10.64YY89 pKa = 9.21MISSEE94 pKa = 4.06CGEE97 pKa = 4.44GKK99 pKa = 10.4GKK101 pKa = 10.25RR102 pKa = 11.84GYY104 pKa = 9.46GQNPHH109 pKa = 5.02YY110 pKa = 10.6HH111 pKa = 6.85IIFDD115 pKa = 4.7LRR117 pKa = 11.84PLVPGASICPIEE129 pKa = 4.34FRR131 pKa = 11.84SLLRR135 pKa = 11.84EE136 pKa = 3.68YY137 pKa = 9.56WQGVDD142 pKa = 4.49EE143 pKa = 5.77DD144 pKa = 4.52PLAPGSKK151 pKa = 8.33PQQYY155 pKa = 10.33KK156 pKa = 10.53FGIVQEE162 pKa = 4.35GQNLGQIDD170 pKa = 3.8NAYY173 pKa = 9.55NASAYY178 pKa = 8.76VSKK181 pKa = 11.09YY182 pKa = 9.68LVKK185 pKa = 10.22TIDD188 pKa = 3.2QRR190 pKa = 11.84KK191 pKa = 7.22EE192 pKa = 3.47ARR194 pKa = 11.84EE195 pKa = 3.91VLRR198 pKa = 11.84EE199 pKa = 4.11LYY201 pKa = 10.41LQVVKK206 pKa = 10.88SFEE209 pKa = 3.88RR210 pKa = 11.84VKK212 pKa = 10.81YY213 pKa = 10.98YY214 pKa = 11.03KK215 pKa = 10.6DD216 pKa = 3.42FLDD219 pKa = 3.77NTFDD223 pKa = 3.48EE224 pKa = 4.89YY225 pKa = 11.59DD226 pKa = 3.58FRR228 pKa = 11.84LHH230 pKa = 7.48RR231 pKa = 11.84YY232 pKa = 6.61YY233 pKa = 10.67QIYY236 pKa = 8.65TEE238 pKa = 4.51TNWCNSPNIPHH249 pKa = 6.72YY250 pKa = 10.77DD251 pKa = 3.17AVPRR255 pKa = 11.84DD256 pKa = 3.99DD257 pKa = 5.03EE258 pKa = 6.27DD259 pKa = 3.89FLEE262 pKa = 4.39WLCLHH267 pKa = 6.45HH268 pKa = 6.46CWNRR272 pKa = 11.84FSEE275 pKa = 4.31WFASDD280 pKa = 2.85INEE283 pKa = 4.36EE284 pKa = 3.88YY285 pKa = 10.43RR286 pKa = 11.84KK287 pKa = 9.98QKK289 pKa = 10.24NEE291 pKa = 3.33YY292 pKa = 8.77RR293 pKa = 11.84NRR295 pKa = 11.84YY296 pKa = 7.52APKK299 pKa = 9.67VRR301 pKa = 11.84ISNGFGDD308 pKa = 3.89YY309 pKa = 11.19ALNFVDD315 pKa = 5.06EE316 pKa = 4.43NNKK319 pKa = 9.02VPLMTFKK326 pKa = 10.66GHH328 pKa = 6.09KK329 pKa = 8.64LKK331 pKa = 10.89KK332 pKa = 9.67IGSHH336 pKa = 7.0LYY338 pKa = 10.0RR339 pKa = 11.84KK340 pKa = 9.75LYY342 pKa = 9.64KK343 pKa = 9.97DD344 pKa = 3.38WYY346 pKa = 9.34VDD348 pKa = 4.3PITKK352 pKa = 8.47TVRR355 pKa = 11.84YY356 pKa = 8.69KK357 pKa = 11.12DD358 pKa = 3.51KK359 pKa = 11.1DD360 pKa = 3.43SAIIGKK366 pKa = 8.49IQTLDD371 pKa = 3.4DD372 pKa = 4.62VILHH376 pKa = 5.86KK377 pKa = 9.51TNCVKK382 pKa = 10.91AKK384 pKa = 9.22LQKK387 pKa = 10.2SSRR390 pKa = 11.84PEE392 pKa = 3.68LASLSNMYY400 pKa = 10.64SIFDD404 pKa = 3.4TVYY407 pKa = 8.55RR408 pKa = 11.84DD409 pKa = 3.49RR410 pKa = 11.84IIRR413 pKa = 11.84LTDD416 pKa = 2.84EE417 pKa = 4.22RR418 pKa = 11.84FYY420 pKa = 11.43DD421 pKa = 4.13DD422 pKa = 4.23SCITPEE428 pKa = 4.07VYY430 pKa = 9.97QDD432 pKa = 3.48YY433 pKa = 10.7LYY435 pKa = 11.39NCLIDD440 pKa = 3.86AQNFNHH446 pKa = 5.97FRR448 pKa = 11.84DD449 pKa = 3.87VTPKK453 pKa = 10.42SDD455 pKa = 3.6YY456 pKa = 10.5FCQKK460 pKa = 10.11FVDD463 pKa = 4.76GVNPEE468 pKa = 4.16HH469 pKa = 6.64MPFFAPFRR477 pKa = 11.84KK478 pKa = 10.23DD479 pKa = 4.03FEE481 pKa = 6.02DD482 pKa = 3.11ITNILDD488 pKa = 3.75ISCAQQDD495 pKa = 3.76QQKK498 pKa = 9.04YY499 pKa = 7.09VAWEE503 pKa = 4.02KK504 pKa = 10.28QKK506 pKa = 11.01KK507 pKa = 9.77VYY509 pKa = 10.46LFQKK513 pKa = 8.35SQNYY517 pKa = 9.54AIQPQVLQQMRR528 pKa = 11.84KK529 pKa = 7.13SS530 pKa = 3.42
MM1 pKa = 8.06RR2 pKa = 11.84LRR4 pKa = 11.84NRR6 pKa = 11.84ITIPTAFAYY15 pKa = 10.09QSQKK19 pKa = 10.29RR20 pKa = 11.84MEE22 pKa = 4.13YY23 pKa = 9.82TIRR26 pKa = 11.84AYY28 pKa = 10.65YY29 pKa = 9.75QFEE32 pKa = 4.08EE33 pKa = 4.59TKK35 pKa = 10.9LKK37 pKa = 10.77GGQQFFYY44 pKa = 10.55TLTYY48 pKa = 10.9NDD50 pKa = 3.69DD51 pKa = 3.92HH52 pKa = 7.63LPTFLGRR59 pKa = 11.84NCHH62 pKa = 6.96DD63 pKa = 4.04YY64 pKa = 11.75NDD66 pKa = 3.44IRR68 pKa = 11.84YY69 pKa = 7.86LTVGAFAKK77 pKa = 10.56KK78 pKa = 10.2LLRR81 pKa = 11.84HH82 pKa = 6.08YY83 pKa = 9.27NLSMKK88 pKa = 10.64YY89 pKa = 9.21MISSEE94 pKa = 4.06CGEE97 pKa = 4.44GKK99 pKa = 10.4GKK101 pKa = 10.25RR102 pKa = 11.84GYY104 pKa = 9.46GQNPHH109 pKa = 5.02YY110 pKa = 10.6HH111 pKa = 6.85IIFDD115 pKa = 4.7LRR117 pKa = 11.84PLVPGASICPIEE129 pKa = 4.34FRR131 pKa = 11.84SLLRR135 pKa = 11.84EE136 pKa = 3.68YY137 pKa = 9.56WQGVDD142 pKa = 4.49EE143 pKa = 5.77DD144 pKa = 4.52PLAPGSKK151 pKa = 8.33PQQYY155 pKa = 10.33KK156 pKa = 10.53FGIVQEE162 pKa = 4.35GQNLGQIDD170 pKa = 3.8NAYY173 pKa = 9.55NASAYY178 pKa = 8.76VSKK181 pKa = 11.09YY182 pKa = 9.68LVKK185 pKa = 10.22TIDD188 pKa = 3.2QRR190 pKa = 11.84KK191 pKa = 7.22EE192 pKa = 3.47ARR194 pKa = 11.84EE195 pKa = 3.91VLRR198 pKa = 11.84EE199 pKa = 4.11LYY201 pKa = 10.41LQVVKK206 pKa = 10.88SFEE209 pKa = 3.88RR210 pKa = 11.84VKK212 pKa = 10.81YY213 pKa = 10.98YY214 pKa = 11.03KK215 pKa = 10.6DD216 pKa = 3.42FLDD219 pKa = 3.77NTFDD223 pKa = 3.48EE224 pKa = 4.89YY225 pKa = 11.59DD226 pKa = 3.58FRR228 pKa = 11.84LHH230 pKa = 7.48RR231 pKa = 11.84YY232 pKa = 6.61YY233 pKa = 10.67QIYY236 pKa = 8.65TEE238 pKa = 4.51TNWCNSPNIPHH249 pKa = 6.72YY250 pKa = 10.77DD251 pKa = 3.17AVPRR255 pKa = 11.84DD256 pKa = 3.99DD257 pKa = 5.03EE258 pKa = 6.27DD259 pKa = 3.89FLEE262 pKa = 4.39WLCLHH267 pKa = 6.45HH268 pKa = 6.46CWNRR272 pKa = 11.84FSEE275 pKa = 4.31WFASDD280 pKa = 2.85INEE283 pKa = 4.36EE284 pKa = 3.88YY285 pKa = 10.43RR286 pKa = 11.84KK287 pKa = 9.98QKK289 pKa = 10.24NEE291 pKa = 3.33YY292 pKa = 8.77RR293 pKa = 11.84NRR295 pKa = 11.84YY296 pKa = 7.52APKK299 pKa = 9.67VRR301 pKa = 11.84ISNGFGDD308 pKa = 3.89YY309 pKa = 11.19ALNFVDD315 pKa = 5.06EE316 pKa = 4.43NNKK319 pKa = 9.02VPLMTFKK326 pKa = 10.66GHH328 pKa = 6.09KK329 pKa = 8.64LKK331 pKa = 10.89KK332 pKa = 9.67IGSHH336 pKa = 7.0LYY338 pKa = 10.0RR339 pKa = 11.84KK340 pKa = 9.75LYY342 pKa = 9.64KK343 pKa = 9.97DD344 pKa = 3.38WYY346 pKa = 9.34VDD348 pKa = 4.3PITKK352 pKa = 8.47TVRR355 pKa = 11.84YY356 pKa = 8.69KK357 pKa = 11.12DD358 pKa = 3.51KK359 pKa = 11.1DD360 pKa = 3.43SAIIGKK366 pKa = 8.49IQTLDD371 pKa = 3.4DD372 pKa = 4.62VILHH376 pKa = 5.86KK377 pKa = 9.51TNCVKK382 pKa = 10.91AKK384 pKa = 9.22LQKK387 pKa = 10.2SSRR390 pKa = 11.84PEE392 pKa = 3.68LASLSNMYY400 pKa = 10.64SIFDD404 pKa = 3.4TVYY407 pKa = 8.55RR408 pKa = 11.84DD409 pKa = 3.49RR410 pKa = 11.84IIRR413 pKa = 11.84LTDD416 pKa = 2.84EE417 pKa = 4.22RR418 pKa = 11.84FYY420 pKa = 11.43DD421 pKa = 4.13DD422 pKa = 4.23SCITPEE428 pKa = 4.07VYY430 pKa = 9.97QDD432 pKa = 3.48YY433 pKa = 10.7LYY435 pKa = 11.39NCLIDD440 pKa = 3.86AQNFNHH446 pKa = 5.97FRR448 pKa = 11.84DD449 pKa = 3.87VTPKK453 pKa = 10.42SDD455 pKa = 3.6YY456 pKa = 10.5FCQKK460 pKa = 10.11FVDD463 pKa = 4.76GVNPEE468 pKa = 4.16HH469 pKa = 6.64MPFFAPFRR477 pKa = 11.84KK478 pKa = 10.23DD479 pKa = 4.03FEE481 pKa = 6.02DD482 pKa = 3.11ITNILDD488 pKa = 3.75ISCAQQDD495 pKa = 3.76QQKK498 pKa = 9.04YY499 pKa = 7.09VAWEE503 pKa = 4.02KK504 pKa = 10.28QKK506 pKa = 11.01KK507 pKa = 9.77VYY509 pKa = 10.46LFQKK513 pKa = 8.35SQNYY517 pKa = 9.54AIQPQVLQQMRR528 pKa = 11.84KK529 pKa = 7.13SS530 pKa = 3.42
Molecular weight: 63.38 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1572 |
98 |
582 |
393.0 |
45.3 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.043 ± 0.913 | 1.209 ± 0.389 |
6.552 ± 0.783 | 6.17 ± 1.08 |
5.344 ± 1.396 | 5.598 ± 0.878 |
1.654 ± 0.403 | 5.725 ± 0.607 |
6.997 ± 0.87 | 8.079 ± 0.5 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.163 ± 0.289 | 6.87 ± 0.67 |
3.69 ± 0.522 | 5.471 ± 0.757 |
5.089 ± 0.516 | 6.743 ± 0.762 |
4.517 ± 0.465 | 5.216 ± 0.205 |
0.954 ± 0.146 | 5.916 ± 1.101 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
