
Lake Sarah-associated circular virus-17
Taxonomy: Viruses; unclassified viruses
Average proteome isoelectric point is 7.15
Get precalculated fractions of proteins

Virtual 2D-PAGE plot for 2 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
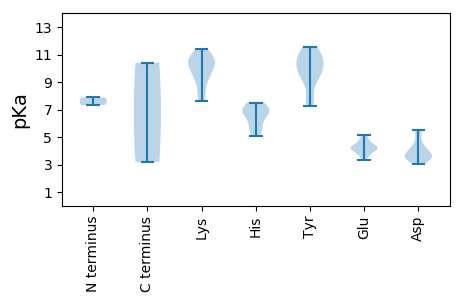
Summary statistics related to proteome-wise predictions


Protein with the lowest isoelectric point:
>tr|A0A140AQL3|A0A140AQL3_9VIRU Coat protein OS=Lake Sarah-associated circular virus-17 OX=1685743 PE=4 SV=1
MM1 pKa = 7.89ANPTRR6 pKa = 11.84QNARR10 pKa = 11.84HH11 pKa = 6.17WIVTQHH17 pKa = 7.19DD18 pKa = 3.35IDD20 pKa = 4.2AFWRR24 pKa = 11.84GDD26 pKa = 3.61PTGHH30 pKa = 6.82HH31 pKa = 6.74AIKK34 pKa = 10.8YY35 pKa = 9.58LVFQVEE41 pKa = 4.34QGAGTSAWHH50 pKa = 5.47IQGFIQFKK58 pKa = 7.61TCVRR62 pKa = 11.84GSVVQRR68 pKa = 11.84LFGGNKK74 pKa = 7.78PHH76 pKa = 7.5VEE78 pKa = 4.36VAHH81 pKa = 6.58HH82 pKa = 6.6PSQARR87 pKa = 11.84EE88 pKa = 3.82YY89 pKa = 10.9CMKK92 pKa = 10.33EE93 pKa = 3.98DD94 pKa = 3.89TQVTPPEE101 pKa = 4.18EE102 pKa = 4.24FGTWEE107 pKa = 3.7PTFAQGYY114 pKa = 9.45RR115 pKa = 11.84SDD117 pKa = 3.67LTVAKK122 pKa = 10.25LQIRR126 pKa = 11.84QHH128 pKa = 5.09GNYY131 pKa = 9.11RR132 pKa = 11.84RR133 pKa = 11.84CLDD136 pKa = 4.36DD137 pKa = 5.54DD138 pKa = 4.68ALDD141 pKa = 4.03TVTARR146 pKa = 11.84YY147 pKa = 7.25PKK149 pKa = 9.67WVADD153 pKa = 3.73QLTMVPRR160 pKa = 11.84PLRR163 pKa = 11.84PIPIVTVYY171 pKa = 10.55YY172 pKa = 10.93GPTNTGKK179 pKa = 8.12TARR182 pKa = 11.84CHH184 pKa = 5.77INNPGIHH191 pKa = 7.34EE192 pKa = 4.08IRR194 pKa = 11.84LDD196 pKa = 3.55NGFINYY202 pKa = 7.76TGQSCVLFDD211 pKa = 5.44EE212 pKa = 4.98FDD214 pKa = 3.65KK215 pKa = 11.42DD216 pKa = 3.31PWPFGTMLKK225 pKa = 10.73LLDD228 pKa = 4.57RR229 pKa = 11.84YY230 pKa = 9.83PFQINVKK237 pKa = 10.1NGYY240 pKa = 7.66SWWEE244 pKa = 3.64ATHH247 pKa = 6.97IFITATEE254 pKa = 4.71APCDD258 pKa = 3.37WYY260 pKa = 11.11LGKK263 pKa = 10.71KK264 pKa = 9.02GVNSDD269 pKa = 5.31HH270 pKa = 6.89IPQFLRR276 pKa = 11.84RR277 pKa = 11.84LTNVVNTTGSVLPPPLPLSPPTLPVPPSPEE307 pKa = 3.35IEE309 pKa = 4.25EE310 pKa = 4.18QQEE313 pKa = 4.1EE314 pKa = 4.51ATMEE318 pKa = 4.5LDD320 pKa = 4.96DD321 pKa = 4.16PTNPEE326 pKa = 3.98DD327 pKa = 3.87YY328 pKa = 10.34EE329 pKa = 4.36NSEE332 pKa = 3.98EE333 pKa = 4.24RR334 pKa = 11.84DD335 pKa = 3.34LRR337 pKa = 11.84EE338 pKa = 3.58EE339 pKa = 3.78MEE341 pKa = 5.14YY342 pKa = 10.67RR343 pKa = 11.84DD344 pKa = 3.9QYY346 pKa = 11.39QGDD349 pKa = 4.01SEE351 pKa = 4.88EE352 pKa = 4.39YY353 pKa = 10.4
MM1 pKa = 7.89ANPTRR6 pKa = 11.84QNARR10 pKa = 11.84HH11 pKa = 6.17WIVTQHH17 pKa = 7.19DD18 pKa = 3.35IDD20 pKa = 4.2AFWRR24 pKa = 11.84GDD26 pKa = 3.61PTGHH30 pKa = 6.82HH31 pKa = 6.74AIKK34 pKa = 10.8YY35 pKa = 9.58LVFQVEE41 pKa = 4.34QGAGTSAWHH50 pKa = 5.47IQGFIQFKK58 pKa = 7.61TCVRR62 pKa = 11.84GSVVQRR68 pKa = 11.84LFGGNKK74 pKa = 7.78PHH76 pKa = 7.5VEE78 pKa = 4.36VAHH81 pKa = 6.58HH82 pKa = 6.6PSQARR87 pKa = 11.84EE88 pKa = 3.82YY89 pKa = 10.9CMKK92 pKa = 10.33EE93 pKa = 3.98DD94 pKa = 3.89TQVTPPEE101 pKa = 4.18EE102 pKa = 4.24FGTWEE107 pKa = 3.7PTFAQGYY114 pKa = 9.45RR115 pKa = 11.84SDD117 pKa = 3.67LTVAKK122 pKa = 10.25LQIRR126 pKa = 11.84QHH128 pKa = 5.09GNYY131 pKa = 9.11RR132 pKa = 11.84RR133 pKa = 11.84CLDD136 pKa = 4.36DD137 pKa = 5.54DD138 pKa = 4.68ALDD141 pKa = 4.03TVTARR146 pKa = 11.84YY147 pKa = 7.25PKK149 pKa = 9.67WVADD153 pKa = 3.73QLTMVPRR160 pKa = 11.84PLRR163 pKa = 11.84PIPIVTVYY171 pKa = 10.55YY172 pKa = 10.93GPTNTGKK179 pKa = 8.12TARR182 pKa = 11.84CHH184 pKa = 5.77INNPGIHH191 pKa = 7.34EE192 pKa = 4.08IRR194 pKa = 11.84LDD196 pKa = 3.55NGFINYY202 pKa = 7.76TGQSCVLFDD211 pKa = 5.44EE212 pKa = 4.98FDD214 pKa = 3.65KK215 pKa = 11.42DD216 pKa = 3.31PWPFGTMLKK225 pKa = 10.73LLDD228 pKa = 4.57RR229 pKa = 11.84YY230 pKa = 9.83PFQINVKK237 pKa = 10.1NGYY240 pKa = 7.66SWWEE244 pKa = 3.64ATHH247 pKa = 6.97IFITATEE254 pKa = 4.71APCDD258 pKa = 3.37WYY260 pKa = 11.11LGKK263 pKa = 10.71KK264 pKa = 9.02GVNSDD269 pKa = 5.31HH270 pKa = 6.89IPQFLRR276 pKa = 11.84RR277 pKa = 11.84LTNVVNTTGSVLPPPLPLSPPTLPVPPSPEE307 pKa = 3.35IEE309 pKa = 4.25EE310 pKa = 4.18QQEE313 pKa = 4.1EE314 pKa = 4.51ATMEE318 pKa = 4.5LDD320 pKa = 4.96DD321 pKa = 4.16PTNPEE326 pKa = 3.98DD327 pKa = 3.87YY328 pKa = 10.34EE329 pKa = 4.36NSEE332 pKa = 3.98EE333 pKa = 4.24RR334 pKa = 11.84DD335 pKa = 3.34LRR337 pKa = 11.84EE338 pKa = 3.58EE339 pKa = 3.78MEE341 pKa = 5.14YY342 pKa = 10.67RR343 pKa = 11.84DD344 pKa = 3.9QYY346 pKa = 11.39QGDD349 pKa = 4.01SEE351 pKa = 4.88EE352 pKa = 4.39YY353 pKa = 10.4
Molecular weight: 40.62 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A140AQL3|A0A140AQL3_9VIRU Coat protein OS=Lake Sarah-associated circular virus-17 OX=1685743 PE=4 SV=1
MM1 pKa = 7.33KK2 pKa = 10.17RR3 pKa = 11.84RR4 pKa = 11.84YY5 pKa = 9.77SSSNLSKK12 pKa = 10.78PFTPPRR18 pKa = 11.84KK19 pKa = 9.15LARR22 pKa = 11.84TTVSVRR28 pKa = 11.84KK29 pKa = 9.49PMLQRR34 pKa = 11.84TMRR37 pKa = 11.84MTMPRR42 pKa = 11.84TEE44 pKa = 4.3LKK46 pKa = 10.44QVDD49 pKa = 4.2VVNPSGGATQAVFPLNSTPSIIPVNLITTGSSAWNRR85 pKa = 11.84IGRR88 pKa = 11.84KK89 pKa = 9.45VNLHH93 pKa = 5.63SCQIQGFFQVNLTTDD108 pKa = 3.03HH109 pKa = 7.3DD110 pKa = 5.3SNVPSWARR118 pKa = 11.84IMLVYY123 pKa = 10.48DD124 pKa = 4.11RR125 pKa = 11.84QPNGATPNINDD136 pKa = 3.61ILLSQVNSATDD147 pKa = 3.76SNVSTPFSATNMNNRR162 pKa = 11.84DD163 pKa = 3.39RR164 pKa = 11.84FEE166 pKa = 4.82IIRR169 pKa = 11.84DD170 pKa = 3.26KK171 pKa = 10.93RR172 pKa = 11.84FCLGSIPLANDD183 pKa = 3.19GQYY186 pKa = 11.53AVGGFEE192 pKa = 4.62NSFSVNMYY200 pKa = 9.45VRR202 pKa = 11.84LANRR206 pKa = 11.84EE207 pKa = 4.13TCFKK211 pKa = 10.58ADD213 pKa = 3.6SSPGVIGDD221 pKa = 3.63IATGSLLFITFGNVISAEE239 pKa = 4.11NPFDD243 pKa = 3.43FAGTVRR249 pKa = 11.84VRR251 pKa = 11.84YY252 pKa = 10.11ADD254 pKa = 3.34VV255 pKa = 3.19
MM1 pKa = 7.33KK2 pKa = 10.17RR3 pKa = 11.84RR4 pKa = 11.84YY5 pKa = 9.77SSSNLSKK12 pKa = 10.78PFTPPRR18 pKa = 11.84KK19 pKa = 9.15LARR22 pKa = 11.84TTVSVRR28 pKa = 11.84KK29 pKa = 9.49PMLQRR34 pKa = 11.84TMRR37 pKa = 11.84MTMPRR42 pKa = 11.84TEE44 pKa = 4.3LKK46 pKa = 10.44QVDD49 pKa = 4.2VVNPSGGATQAVFPLNSTPSIIPVNLITTGSSAWNRR85 pKa = 11.84IGRR88 pKa = 11.84KK89 pKa = 9.45VNLHH93 pKa = 5.63SCQIQGFFQVNLTTDD108 pKa = 3.03HH109 pKa = 7.3DD110 pKa = 5.3SNVPSWARR118 pKa = 11.84IMLVYY123 pKa = 10.48DD124 pKa = 4.11RR125 pKa = 11.84QPNGATPNINDD136 pKa = 3.61ILLSQVNSATDD147 pKa = 3.76SNVSTPFSATNMNNRR162 pKa = 11.84DD163 pKa = 3.39RR164 pKa = 11.84FEE166 pKa = 4.82IIRR169 pKa = 11.84DD170 pKa = 3.26KK171 pKa = 10.93RR172 pKa = 11.84FCLGSIPLANDD183 pKa = 3.19GQYY186 pKa = 11.53AVGGFEE192 pKa = 4.62NSFSVNMYY200 pKa = 9.45VRR202 pKa = 11.84LANRR206 pKa = 11.84EE207 pKa = 4.13TCFKK211 pKa = 10.58ADD213 pKa = 3.6SSPGVIGDD221 pKa = 3.63IATGSLLFITFGNVISAEE239 pKa = 4.11NPFDD243 pKa = 3.43FAGTVRR249 pKa = 11.84VRR251 pKa = 11.84YY252 pKa = 10.11ADD254 pKa = 3.34VV255 pKa = 3.19
Molecular weight: 28.19 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
608 |
255 |
353 |
304.0 |
34.4 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
5.592 ± 0.42 | 1.48 ± 0.187 |
5.921 ± 0.507 | 5.099 ± 1.932 |
4.605 ± 0.545 | 6.086 ± 0.125 |
2.467 ± 1.036 | 5.263 ± 0.381 |
3.289 ± 0.094 | 6.25 ± 0.015 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.303 ± 0.514 | 6.25 ± 1.464 |
7.73 ± 0.896 | 4.77 ± 0.764 |
6.743 ± 0.677 | 6.086 ± 2.29 |
7.895 ± 0.032 | 7.072 ± 0.716 |
1.809 ± 0.631 | 3.289 ± 0.818 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
