
Wuhan spider virus 5
Taxonomy: Viruses; Riboviria; unclassified Riboviria; unclassified RNA viruses ShiM-2016
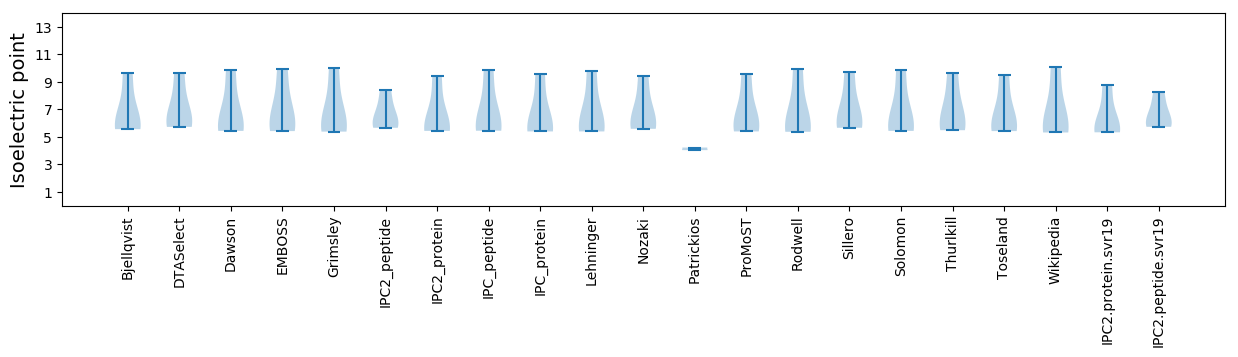
Average proteome isoelectric point is 6.34
Get precalculated fractions of proteins

Virtual 2D-PAGE plot for 4 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions

Protein with the lowest isoelectric point:
>tr|A0A1L3KJ82|A0A1L3KJ82_9VIRU Uncharacterized protein OS=Wuhan spider virus 5 OX=1923754 PE=4 SV=1
MM1 pKa = 7.37ATPIAQANSSVSNGLDD17 pKa = 3.1LPSTVEE23 pKa = 4.2AGAHH27 pKa = 5.8LPSEE31 pKa = 4.59IQTPTPPTVEE41 pKa = 4.09VPFAAGEE48 pKa = 4.21GNRR51 pKa = 11.84LDD53 pKa = 3.68PYY55 pKa = 10.64FYY57 pKa = 10.45EE58 pKa = 3.96QFIKK62 pKa = 11.1VEE64 pKa = 4.08VFPWTTMDD72 pKa = 3.41QPGKK76 pKa = 10.61VLFVLKK82 pKa = 10.56LDD84 pKa = 3.98PKK86 pKa = 10.86NINKK90 pKa = 9.01QLAYY94 pKa = 10.58FLDD97 pKa = 4.85AYY99 pKa = 8.87YY100 pKa = 11.18AWGGDD105 pKa = 2.87IKK107 pKa = 11.22LLLKK111 pKa = 10.86VLGTSFHH118 pKa = 6.83AGQLGIYY125 pKa = 7.56YY126 pKa = 9.38LPPGVDD132 pKa = 3.34YY133 pKa = 11.33KK134 pKa = 11.47NYY136 pKa = 9.3TPQEE140 pKa = 3.76LSIFPWEE147 pKa = 4.07IVDD150 pKa = 3.73VKK152 pKa = 10.84DD153 pKa = 3.9INLHH157 pKa = 5.91EE158 pKa = 4.32ILMRR162 pKa = 11.84DD163 pKa = 3.13IRR165 pKa = 11.84PTKK168 pKa = 10.43YY169 pKa = 10.37HH170 pKa = 5.75MLEE173 pKa = 4.88DD174 pKa = 3.88KK175 pKa = 10.96PNDD178 pKa = 3.51PFYY181 pKa = 10.33VQSGGTLVIAADD193 pKa = 4.13TQLSTSSTGVNKK205 pKa = 10.02IYY207 pKa = 10.85VSVWSKK213 pKa = 10.39LAEE216 pKa = 4.26NFQVAYY222 pKa = 8.11MIPPRR227 pKa = 11.84EE228 pKa = 4.36LKK230 pKa = 10.54PSLNFSNEE238 pKa = 2.79FMAAVNSIFYY248 pKa = 9.63TGSKK252 pKa = 9.98SLASAPFRR260 pKa = 11.84IDD262 pKa = 3.3TIDD265 pKa = 3.47IQPVTIRR272 pKa = 11.84QTFNGIYY279 pKa = 10.4NCFTTAGEE287 pKa = 4.1NLYY290 pKa = 9.78TPILPDD296 pKa = 4.63DD297 pKa = 4.07YY298 pKa = 11.27PNSKK302 pKa = 10.25FNLPIYY308 pKa = 10.19PMAGTFRR315 pKa = 11.84TTSNPFEE322 pKa = 4.18VEE324 pKa = 3.39ISFGFWEE331 pKa = 5.2FWPKK335 pKa = 10.74EE336 pKa = 3.75FTISYY341 pKa = 10.37KK342 pKa = 11.01DD343 pKa = 3.26GDD345 pKa = 4.11KK346 pKa = 10.21RR347 pKa = 11.84TSATPSDD354 pKa = 3.92MSWDD358 pKa = 3.28KK359 pKa = 11.52SDD361 pKa = 3.54PFKK364 pKa = 11.44VSFTHH369 pKa = 7.48EE370 pKa = 4.77DD371 pKa = 3.63GLSVKK376 pKa = 9.73IDD378 pKa = 3.55EE379 pKa = 4.59PVKK382 pKa = 10.45FFPDD386 pKa = 3.28NVGVFTDD393 pKa = 3.78KK394 pKa = 10.89VSPVNQSTTPVNTPQARR411 pKa = 11.84EE412 pKa = 4.1SIVYY416 pKa = 7.68FQNSFGAYY424 pKa = 8.49SLQPAVLAQACADD437 pKa = 3.58KK438 pKa = 11.18QFIDD442 pKa = 4.96VIPQGMAALLIMTEE456 pKa = 4.18KK457 pKa = 10.45TSGLPLTYY465 pKa = 10.5LKK467 pKa = 10.44FYY469 pKa = 10.84NRR471 pKa = 11.84GIFTAPASVDD481 pKa = 3.64LVKK484 pKa = 10.7FSLDD488 pKa = 3.24DD489 pKa = 3.77KK490 pKa = 11.1QIKK493 pKa = 9.8FSNFVLEE500 pKa = 4.49TQSFPVNEE508 pKa = 3.84EE509 pKa = 3.8MIRR512 pKa = 11.84NMLAYY517 pKa = 10.5CSFHH521 pKa = 5.72TNKK524 pKa = 10.53KK525 pKa = 7.78IMRR528 pKa = 11.84RR529 pKa = 11.84ISSAKK534 pKa = 9.72SLQSRR539 pKa = 11.84KK540 pKa = 9.55ALKK543 pKa = 10.57KK544 pKa = 9.99FVTFQEE550 pKa = 4.32TGII553 pKa = 4.1
MM1 pKa = 7.37ATPIAQANSSVSNGLDD17 pKa = 3.1LPSTVEE23 pKa = 4.2AGAHH27 pKa = 5.8LPSEE31 pKa = 4.59IQTPTPPTVEE41 pKa = 4.09VPFAAGEE48 pKa = 4.21GNRR51 pKa = 11.84LDD53 pKa = 3.68PYY55 pKa = 10.64FYY57 pKa = 10.45EE58 pKa = 3.96QFIKK62 pKa = 11.1VEE64 pKa = 4.08VFPWTTMDD72 pKa = 3.41QPGKK76 pKa = 10.61VLFVLKK82 pKa = 10.56LDD84 pKa = 3.98PKK86 pKa = 10.86NINKK90 pKa = 9.01QLAYY94 pKa = 10.58FLDD97 pKa = 4.85AYY99 pKa = 8.87YY100 pKa = 11.18AWGGDD105 pKa = 2.87IKK107 pKa = 11.22LLLKK111 pKa = 10.86VLGTSFHH118 pKa = 6.83AGQLGIYY125 pKa = 7.56YY126 pKa = 9.38LPPGVDD132 pKa = 3.34YY133 pKa = 11.33KK134 pKa = 11.47NYY136 pKa = 9.3TPQEE140 pKa = 3.76LSIFPWEE147 pKa = 4.07IVDD150 pKa = 3.73VKK152 pKa = 10.84DD153 pKa = 3.9INLHH157 pKa = 5.91EE158 pKa = 4.32ILMRR162 pKa = 11.84DD163 pKa = 3.13IRR165 pKa = 11.84PTKK168 pKa = 10.43YY169 pKa = 10.37HH170 pKa = 5.75MLEE173 pKa = 4.88DD174 pKa = 3.88KK175 pKa = 10.96PNDD178 pKa = 3.51PFYY181 pKa = 10.33VQSGGTLVIAADD193 pKa = 4.13TQLSTSSTGVNKK205 pKa = 10.02IYY207 pKa = 10.85VSVWSKK213 pKa = 10.39LAEE216 pKa = 4.26NFQVAYY222 pKa = 8.11MIPPRR227 pKa = 11.84EE228 pKa = 4.36LKK230 pKa = 10.54PSLNFSNEE238 pKa = 2.79FMAAVNSIFYY248 pKa = 9.63TGSKK252 pKa = 9.98SLASAPFRR260 pKa = 11.84IDD262 pKa = 3.3TIDD265 pKa = 3.47IQPVTIRR272 pKa = 11.84QTFNGIYY279 pKa = 10.4NCFTTAGEE287 pKa = 4.1NLYY290 pKa = 9.78TPILPDD296 pKa = 4.63DD297 pKa = 4.07YY298 pKa = 11.27PNSKK302 pKa = 10.25FNLPIYY308 pKa = 10.19PMAGTFRR315 pKa = 11.84TTSNPFEE322 pKa = 4.18VEE324 pKa = 3.39ISFGFWEE331 pKa = 5.2FWPKK335 pKa = 10.74EE336 pKa = 3.75FTISYY341 pKa = 10.37KK342 pKa = 11.01DD343 pKa = 3.26GDD345 pKa = 4.11KK346 pKa = 10.21RR347 pKa = 11.84TSATPSDD354 pKa = 3.92MSWDD358 pKa = 3.28KK359 pKa = 11.52SDD361 pKa = 3.54PFKK364 pKa = 11.44VSFTHH369 pKa = 7.48EE370 pKa = 4.77DD371 pKa = 3.63GLSVKK376 pKa = 9.73IDD378 pKa = 3.55EE379 pKa = 4.59PVKK382 pKa = 10.45FFPDD386 pKa = 3.28NVGVFTDD393 pKa = 3.78KK394 pKa = 10.89VSPVNQSTTPVNTPQARR411 pKa = 11.84EE412 pKa = 4.1SIVYY416 pKa = 7.68FQNSFGAYY424 pKa = 8.49SLQPAVLAQACADD437 pKa = 3.58KK438 pKa = 11.18QFIDD442 pKa = 4.96VIPQGMAALLIMTEE456 pKa = 4.18KK457 pKa = 10.45TSGLPLTYY465 pKa = 10.5LKK467 pKa = 10.44FYY469 pKa = 10.84NRR471 pKa = 11.84GIFTAPASVDD481 pKa = 3.64LVKK484 pKa = 10.7FSLDD488 pKa = 3.24DD489 pKa = 3.77KK490 pKa = 11.1QIKK493 pKa = 9.8FSNFVLEE500 pKa = 4.49TQSFPVNEE508 pKa = 3.84EE509 pKa = 3.8MIRR512 pKa = 11.84NMLAYY517 pKa = 10.5CSFHH521 pKa = 5.72TNKK524 pKa = 10.53KK525 pKa = 7.78IMRR528 pKa = 11.84RR529 pKa = 11.84ISSAKK534 pKa = 9.72SLQSRR539 pKa = 11.84KK540 pKa = 9.55ALKK543 pKa = 10.57KK544 pKa = 9.99FVTFQEE550 pKa = 4.32TGII553 pKa = 4.1
Molecular weight: 62.19 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1L3KJ39|A0A1L3KJ39_9VIRU Calici_coat domain-containing protein OS=Wuhan spider virus 5 OX=1923754 PE=4 SV=1
MM1 pKa = 7.47ALVALGAAALAGGISAGATLGASLGAADD29 pKa = 4.37VQSKK33 pKa = 7.79TATSINNQNLDD44 pKa = 3.53FAKK47 pKa = 10.02QQYY50 pKa = 10.35SEE52 pKa = 4.32GLSSFKK58 pKa = 10.51QAGLPSFLYY67 pKa = 10.9YY68 pKa = 10.69SGGNSGLGNLPKK80 pKa = 9.58TQSHH84 pKa = 6.52IEE86 pKa = 4.01GSSFSSSLGVNADD99 pKa = 3.5LPYY102 pKa = 11.11YY103 pKa = 10.09SANPYY108 pKa = 9.02NQYY111 pKa = 10.52LQTGRR116 pKa = 11.84PVPTNKK122 pKa = 8.16QTQEE126 pKa = 4.26RR127 pKa = 11.84IPNSNNNTPRR137 pKa = 11.84SNDD140 pKa = 2.8IEE142 pKa = 4.22LQDD145 pKa = 4.07FSSIEE150 pKa = 3.78PTSRR154 pKa = 11.84ITNSNGNNFQYY165 pKa = 10.86GGSTSMDD172 pKa = 3.27NLRR175 pKa = 11.84AQFAPMLGGNRR186 pKa = 11.84QSGSGKK192 pKa = 9.84YY193 pKa = 10.37SSVPPPASYY202 pKa = 7.93FTSNRR207 pKa = 11.84YY208 pKa = 8.78SQTPPNFGNVSKK220 pKa = 10.87SSQTSYY226 pKa = 11.32SSNFGSNSATNNTRR240 pKa = 11.84SFYY243 pKa = 10.83SQTNGIFFRR252 pKa = 11.84SVGFNPLSARR262 pKa = 11.84GRR264 pKa = 3.68
MM1 pKa = 7.47ALVALGAAALAGGISAGATLGASLGAADD29 pKa = 4.37VQSKK33 pKa = 7.79TATSINNQNLDD44 pKa = 3.53FAKK47 pKa = 10.02QQYY50 pKa = 10.35SEE52 pKa = 4.32GLSSFKK58 pKa = 10.51QAGLPSFLYY67 pKa = 10.9YY68 pKa = 10.69SGGNSGLGNLPKK80 pKa = 9.58TQSHH84 pKa = 6.52IEE86 pKa = 4.01GSSFSSSLGVNADD99 pKa = 3.5LPYY102 pKa = 11.11YY103 pKa = 10.09SANPYY108 pKa = 9.02NQYY111 pKa = 10.52LQTGRR116 pKa = 11.84PVPTNKK122 pKa = 8.16QTQEE126 pKa = 4.26RR127 pKa = 11.84IPNSNNNTPRR137 pKa = 11.84SNDD140 pKa = 2.8IEE142 pKa = 4.22LQDD145 pKa = 4.07FSSIEE150 pKa = 3.78PTSRR154 pKa = 11.84ITNSNGNNFQYY165 pKa = 10.86GGSTSMDD172 pKa = 3.27NLRR175 pKa = 11.84AQFAPMLGGNRR186 pKa = 11.84QSGSGKK192 pKa = 9.84YY193 pKa = 10.37SSVPPPASYY202 pKa = 7.93FTSNRR207 pKa = 11.84YY208 pKa = 8.78SQTPPNFGNVSKK220 pKa = 10.87SSQTSYY226 pKa = 11.32SSNFGSNSATNNTRR240 pKa = 11.84SFYY243 pKa = 10.83SQTNGIFFRR252 pKa = 11.84SVGFNPLSARR262 pKa = 11.84GRR264 pKa = 3.68
Molecular weight: 27.94 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
3565 |
218 |
2530 |
891.3 |
100.94 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
5.61 ± 0.792 | 1.683 ± 0.463 |
5.75 ± 0.621 | 5.891 ± 0.717 |
5.217 ± 0.54 | 4.769 ± 0.822 |
2.328 ± 0.674 | 6.396 ± 0.819 |
6.76 ± 0.68 | 8.387 ± 0.34 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.384 ± 0.183 | 5.806 ± 0.853 |
5.61 ± 0.581 | 3.506 ± 0.682 |
4.039 ± 0.392 | 8.527 ± 1.314 |
5.666 ± 0.446 | 6.424 ± 0.655 |
0.954 ± 0.209 | 4.292 ± 0.14 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
