
Tortoise microvirus 19
Taxonomy: Viruses; Monodnaviria; Sangervirae; Phixviricota; Malgrandaviricetes; Petitvirales; Microviridae; unclassified Microviridae
Average proteome isoelectric point is 6.58
Get precalculated fractions of proteins
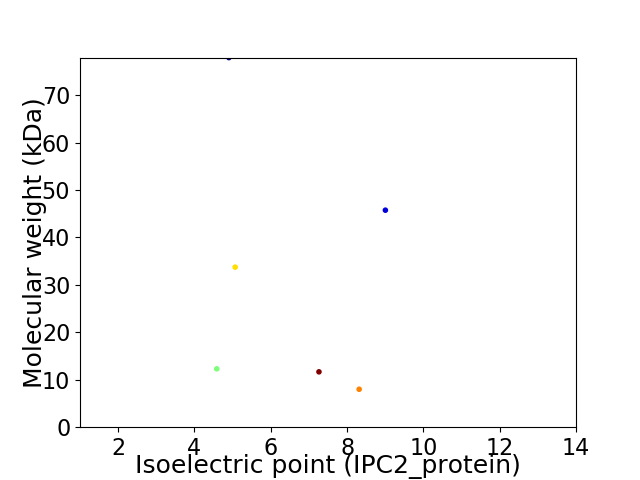
Virtual 2D-PAGE plot for 6 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A4P8W6X5|A0A4P8W6X5_9VIRU Replication initiation protein OS=Tortoise microvirus 19 OX=2583120 PE=4 SV=1
MM1 pKa = 7.77EE2 pKa = 5.19EE3 pKa = 4.07NEE5 pKa = 4.35KK6 pKa = 10.91QNSLGLSQDD15 pKa = 2.93KK16 pKa = 11.1ANYY19 pKa = 9.37SDD21 pKa = 3.7EE22 pKa = 4.23EE23 pKa = 4.53RR24 pKa = 11.84IEE26 pKa = 3.97MQRR29 pKa = 11.84GIVGTPLKK37 pKa = 10.27VVRR40 pKa = 11.84VEE42 pKa = 3.92EE43 pKa = 4.34TYY45 pKa = 10.64KK46 pKa = 10.52IAFGKK51 pKa = 10.86YY52 pKa = 8.28IVCSKK57 pKa = 10.95NFGTIEE63 pKa = 3.76EE64 pKa = 4.32AEE66 pKa = 4.4EE67 pKa = 4.02FVEE70 pKa = 4.65TKK72 pKa = 10.27PGLWDD77 pKa = 3.25IVVGLMGVATTEE89 pKa = 4.37AIKK92 pKa = 10.45EE93 pKa = 4.01ISRR96 pKa = 11.84EE97 pKa = 3.96LTDD100 pKa = 3.61EE101 pKa = 4.25LKK103 pKa = 10.76EE104 pKa = 4.06ITGEE108 pKa = 4.01QQ109 pKa = 3.26
MM1 pKa = 7.77EE2 pKa = 5.19EE3 pKa = 4.07NEE5 pKa = 4.35KK6 pKa = 10.91QNSLGLSQDD15 pKa = 2.93KK16 pKa = 11.1ANYY19 pKa = 9.37SDD21 pKa = 3.7EE22 pKa = 4.23EE23 pKa = 4.53RR24 pKa = 11.84IEE26 pKa = 3.97MQRR29 pKa = 11.84GIVGTPLKK37 pKa = 10.27VVRR40 pKa = 11.84VEE42 pKa = 3.92EE43 pKa = 4.34TYY45 pKa = 10.64KK46 pKa = 10.52IAFGKK51 pKa = 10.86YY52 pKa = 8.28IVCSKK57 pKa = 10.95NFGTIEE63 pKa = 3.76EE64 pKa = 4.32AEE66 pKa = 4.4EE67 pKa = 4.02FVEE70 pKa = 4.65TKK72 pKa = 10.27PGLWDD77 pKa = 3.25IVVGLMGVATTEE89 pKa = 4.37AIKK92 pKa = 10.45EE93 pKa = 4.01ISRR96 pKa = 11.84EE97 pKa = 3.96LTDD100 pKa = 3.61EE101 pKa = 4.25LKK103 pKa = 10.76EE104 pKa = 4.06ITGEE108 pKa = 4.01QQ109 pKa = 3.26
Molecular weight: 12.3 kDa
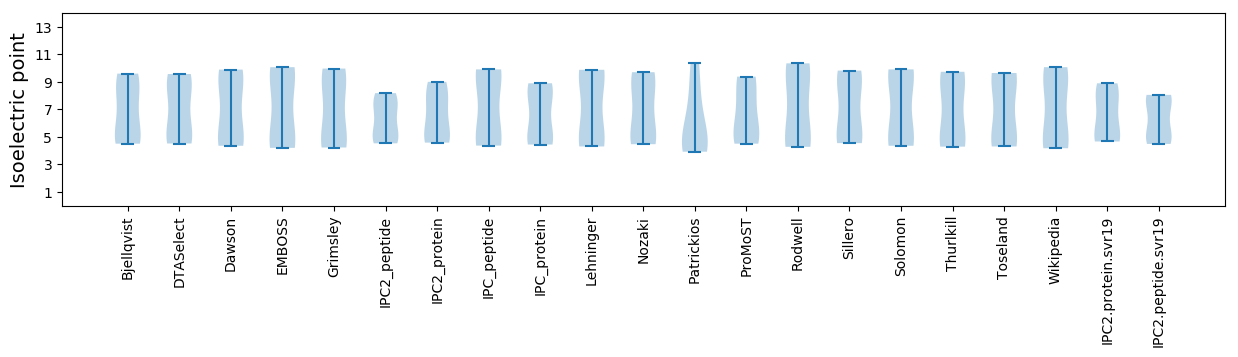
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A4P8W9A8|A0A4P8W9A8_9VIRU DNA pilot protein OS=Tortoise microvirus 19 OX=2583120 PE=4 SV=1
MM1 pKa = 7.56CLYY4 pKa = 10.5TKK6 pKa = 9.98MVLNPKK12 pKa = 9.57YY13 pKa = 10.42KK14 pKa = 9.82PSKK17 pKa = 10.22KK18 pKa = 10.13NGGNPPVCTDD28 pKa = 3.22EE29 pKa = 4.01RR30 pKa = 11.84TKK32 pKa = 10.82YY33 pKa = 10.59IPVSCGNCIEE43 pKa = 4.2CRR45 pKa = 11.84KK46 pKa = 9.74KK47 pKa = 10.37KK48 pKa = 8.88QRR50 pKa = 11.84EE51 pKa = 3.67WRR53 pKa = 11.84VRR55 pKa = 11.84MLEE58 pKa = 4.12EE59 pKa = 4.1IKK61 pKa = 10.36HH62 pKa = 6.02DD63 pKa = 3.58KK64 pKa = 9.18TGKK67 pKa = 10.01FVTLTFSEE75 pKa = 4.42EE76 pKa = 3.74ALIEE80 pKa = 4.05LEE82 pKa = 4.11IEE84 pKa = 4.45AGTKK88 pKa = 8.47EE89 pKa = 4.37ANTVATLAVRR99 pKa = 11.84RR100 pKa = 11.84FLEE103 pKa = 4.1RR104 pKa = 11.84WRR106 pKa = 11.84KK107 pKa = 8.56EE108 pKa = 3.4YY109 pKa = 10.54RR110 pKa = 11.84KK111 pKa = 9.81SVKK114 pKa = 9.48HH115 pKa = 4.99WFITEE120 pKa = 4.24LGHH123 pKa = 7.29RR124 pKa = 11.84NTEE127 pKa = 3.82RR128 pKa = 11.84VHH130 pKa = 6.01IHH132 pKa = 6.73GIIFTDD138 pKa = 3.87KK139 pKa = 10.46EE140 pKa = 4.14KK141 pKa = 11.27EE142 pKa = 3.99DD143 pKa = 3.43IQRR146 pKa = 11.84IWKK149 pKa = 9.67YY150 pKa = 10.57GRR152 pKa = 11.84TDD154 pKa = 2.68TGYY157 pKa = 11.67SMGEE161 pKa = 3.65RR162 pKa = 11.84CVNYY166 pKa = 9.84VMKK169 pKa = 10.73YY170 pKa = 6.82ITKK173 pKa = 9.44TDD175 pKa = 3.38QDD177 pKa = 3.47HH178 pKa = 6.65KK179 pKa = 11.23GFVGKK184 pKa = 10.2ILTSPGIGRR193 pKa = 11.84GYY195 pKa = 9.59IGSYY199 pKa = 9.57NASKK203 pKa = 11.19NKK205 pKa = 9.52FNKK208 pKa = 9.78EE209 pKa = 4.18STNEE213 pKa = 4.01TYY215 pKa = 11.05KK216 pKa = 11.14LNNGCKK222 pKa = 9.26IALPQYY228 pKa = 9.31YY229 pKa = 9.54RR230 pKa = 11.84NKK232 pKa = 10.18IYY234 pKa = 10.51TEE236 pKa = 4.13EE237 pKa = 4.21EE238 pKa = 4.37KK239 pKa = 10.85EE240 pKa = 3.91QLWISKK246 pKa = 9.78IEE248 pKa = 3.97KK249 pKa = 9.68EE250 pKa = 4.16EE251 pKa = 4.46LYY253 pKa = 11.58VMGRR257 pKa = 11.84KK258 pKa = 8.67IRR260 pKa = 11.84NINTEE265 pKa = 3.69RR266 pKa = 11.84GQRR269 pKa = 11.84EE270 pKa = 4.08LKK272 pKa = 10.64YY273 pKa = 10.66AIEE276 pKa = 4.08YY277 pKa = 9.5AKK279 pKa = 10.48KK280 pKa = 9.8ISQQQGYY287 pKa = 8.5GAGEE291 pKa = 4.08GKK293 pKa = 9.71KK294 pKa = 10.61AYY296 pKa = 7.08MTRR299 pKa = 11.84KK300 pKa = 10.12NGTKK304 pKa = 9.28YY305 pKa = 10.88LEE307 pKa = 4.18VNRR310 pKa = 11.84RR311 pKa = 11.84IHH313 pKa = 6.71NIATMFEE320 pKa = 3.96EE321 pKa = 4.47SEE323 pKa = 4.32RR324 pKa = 11.84VAKK327 pKa = 10.6ARR329 pKa = 11.84IKK331 pKa = 10.05HH332 pKa = 5.1QNSINYY338 pKa = 8.23LNSMSYY344 pKa = 10.71EE345 pKa = 3.63EE346 pKa = 4.64SIRR349 pKa = 11.84NFSEE353 pKa = 3.57NEE355 pKa = 3.58HH356 pKa = 6.15RR357 pKa = 11.84VNSTGRR363 pKa = 11.84NIPAEE368 pKa = 3.95NEE370 pKa = 3.9GRR372 pKa = 11.84KK373 pKa = 9.82DD374 pKa = 3.45EE375 pKa = 4.62PGKK378 pKa = 9.36RR379 pKa = 11.84RR380 pKa = 11.84SYY382 pKa = 10.49RR383 pKa = 11.84GHH385 pKa = 6.43EE386 pKa = 3.96KK387 pKa = 10.79DD388 pKa = 3.35GG389 pKa = 3.74
MM1 pKa = 7.56CLYY4 pKa = 10.5TKK6 pKa = 9.98MVLNPKK12 pKa = 9.57YY13 pKa = 10.42KK14 pKa = 9.82PSKK17 pKa = 10.22KK18 pKa = 10.13NGGNPPVCTDD28 pKa = 3.22EE29 pKa = 4.01RR30 pKa = 11.84TKK32 pKa = 10.82YY33 pKa = 10.59IPVSCGNCIEE43 pKa = 4.2CRR45 pKa = 11.84KK46 pKa = 9.74KK47 pKa = 10.37KK48 pKa = 8.88QRR50 pKa = 11.84EE51 pKa = 3.67WRR53 pKa = 11.84VRR55 pKa = 11.84MLEE58 pKa = 4.12EE59 pKa = 4.1IKK61 pKa = 10.36HH62 pKa = 6.02DD63 pKa = 3.58KK64 pKa = 9.18TGKK67 pKa = 10.01FVTLTFSEE75 pKa = 4.42EE76 pKa = 3.74ALIEE80 pKa = 4.05LEE82 pKa = 4.11IEE84 pKa = 4.45AGTKK88 pKa = 8.47EE89 pKa = 4.37ANTVATLAVRR99 pKa = 11.84RR100 pKa = 11.84FLEE103 pKa = 4.1RR104 pKa = 11.84WRR106 pKa = 11.84KK107 pKa = 8.56EE108 pKa = 3.4YY109 pKa = 10.54RR110 pKa = 11.84KK111 pKa = 9.81SVKK114 pKa = 9.48HH115 pKa = 4.99WFITEE120 pKa = 4.24LGHH123 pKa = 7.29RR124 pKa = 11.84NTEE127 pKa = 3.82RR128 pKa = 11.84VHH130 pKa = 6.01IHH132 pKa = 6.73GIIFTDD138 pKa = 3.87KK139 pKa = 10.46EE140 pKa = 4.14KK141 pKa = 11.27EE142 pKa = 3.99DD143 pKa = 3.43IQRR146 pKa = 11.84IWKK149 pKa = 9.67YY150 pKa = 10.57GRR152 pKa = 11.84TDD154 pKa = 2.68TGYY157 pKa = 11.67SMGEE161 pKa = 3.65RR162 pKa = 11.84CVNYY166 pKa = 9.84VMKK169 pKa = 10.73YY170 pKa = 6.82ITKK173 pKa = 9.44TDD175 pKa = 3.38QDD177 pKa = 3.47HH178 pKa = 6.65KK179 pKa = 11.23GFVGKK184 pKa = 10.2ILTSPGIGRR193 pKa = 11.84GYY195 pKa = 9.59IGSYY199 pKa = 9.57NASKK203 pKa = 11.19NKK205 pKa = 9.52FNKK208 pKa = 9.78EE209 pKa = 4.18STNEE213 pKa = 4.01TYY215 pKa = 11.05KK216 pKa = 11.14LNNGCKK222 pKa = 9.26IALPQYY228 pKa = 9.31YY229 pKa = 9.54RR230 pKa = 11.84NKK232 pKa = 10.18IYY234 pKa = 10.51TEE236 pKa = 4.13EE237 pKa = 4.21EE238 pKa = 4.37KK239 pKa = 10.85EE240 pKa = 3.91QLWISKK246 pKa = 9.78IEE248 pKa = 3.97KK249 pKa = 9.68EE250 pKa = 4.16EE251 pKa = 4.46LYY253 pKa = 11.58VMGRR257 pKa = 11.84KK258 pKa = 8.67IRR260 pKa = 11.84NINTEE265 pKa = 3.69RR266 pKa = 11.84GQRR269 pKa = 11.84EE270 pKa = 4.08LKK272 pKa = 10.64YY273 pKa = 10.66AIEE276 pKa = 4.08YY277 pKa = 9.5AKK279 pKa = 10.48KK280 pKa = 9.8ISQQQGYY287 pKa = 8.5GAGEE291 pKa = 4.08GKK293 pKa = 9.71KK294 pKa = 10.61AYY296 pKa = 7.08MTRR299 pKa = 11.84KK300 pKa = 10.12NGTKK304 pKa = 9.28YY305 pKa = 10.88LEE307 pKa = 4.18VNRR310 pKa = 11.84RR311 pKa = 11.84IHH313 pKa = 6.71NIATMFEE320 pKa = 3.96EE321 pKa = 4.47SEE323 pKa = 4.32RR324 pKa = 11.84VAKK327 pKa = 10.6ARR329 pKa = 11.84IKK331 pKa = 10.05HH332 pKa = 5.1QNSINYY338 pKa = 8.23LNSMSYY344 pKa = 10.71EE345 pKa = 3.63EE346 pKa = 4.64SIRR349 pKa = 11.84NFSEE353 pKa = 3.57NEE355 pKa = 3.58HH356 pKa = 6.15RR357 pKa = 11.84VNSTGRR363 pKa = 11.84NIPAEE368 pKa = 3.95NEE370 pKa = 3.9GRR372 pKa = 11.84KK373 pKa = 9.82DD374 pKa = 3.45EE375 pKa = 4.62PGKK378 pKa = 9.36RR379 pKa = 11.84RR380 pKa = 11.84SYY382 pKa = 10.49RR383 pKa = 11.84GHH385 pKa = 6.43EE386 pKa = 3.96KK387 pKa = 10.79DD388 pKa = 3.35GG389 pKa = 3.74
Molecular weight: 45.74 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1685 |
68 |
696 |
280.8 |
31.56 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.3 ± 2.204 | 0.593 ± 0.321 |
4.688 ± 0.887 | 9.614 ± 1.99 |
2.611 ± 0.601 | 8.843 ± 0.778 |
1.543 ± 0.394 | 6.588 ± 0.425 |
7.596 ± 1.658 | 6.231 ± 0.536 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.73 ± 0.544 | 5.46 ± 0.686 |
2.671 ± 0.569 | 4.273 ± 0.729 |
5.697 ± 1.055 | 5.223 ± 0.328 |
7.774 ± 1.155 | 5.223 ± 0.595 |
1.602 ± 0.302 | 3.739 ± 0.817 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
