
Gossypium barbadense (Sea-island cotton) (Egyptian cotton)
Taxonomy: cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliopsida; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; malvids; Malvales; Malvaceae; Malvoideae; Gossypium
Average proteome isoelectric point is 6.81
Get precalculated fractions of proteins
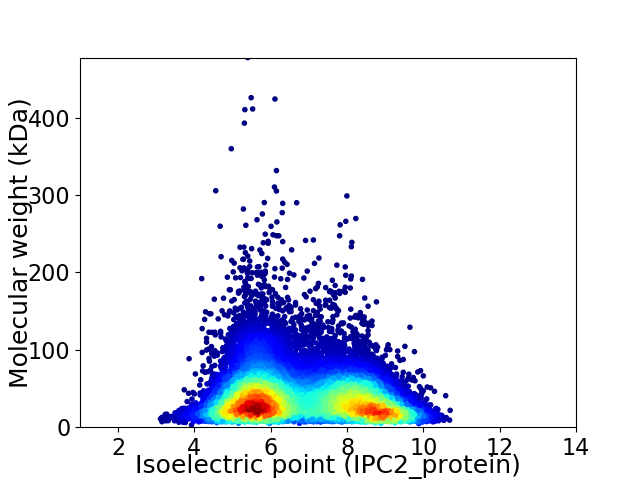
Virtual 2D-PAGE plot for 40046 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A2P5XLG9|A0A2P5XLG9_GOSBA Uncharacterized protein OS=Gossypium barbadense OX=3634 GN=GOBAR_AA16507 PE=4 SV=1
MM1 pKa = 7.51IALYY5 pKa = 10.6CEE7 pKa = 4.05NRR9 pKa = 11.84SDD11 pKa = 3.51KK12 pKa = 10.65YY13 pKa = 11.56APIHH17 pKa = 6.41LFAEE21 pKa = 4.49LAGMKK26 pKa = 10.12QNEE29 pKa = 4.59DD30 pKa = 2.96FTAYY34 pKa = 10.38GEE36 pKa = 3.99EE37 pKa = 4.01HH38 pKa = 6.97RR39 pKa = 11.84AQEE42 pKa = 4.68LCMMAPISYY51 pKa = 10.13VDD53 pKa = 3.44SEE55 pKa = 4.25LTIRR59 pKa = 11.84GINIDD64 pKa = 4.25LNVTPDD70 pKa = 3.18IDD72 pKa = 4.01VVGDD76 pKa = 3.81DD77 pKa = 5.53GYY79 pKa = 11.52DD80 pKa = 3.37SSDD83 pKa = 3.43PCDD86 pKa = 3.33QKK88 pKa = 11.51VDD90 pKa = 3.71SDD92 pKa = 3.86NDD94 pKa = 3.82LDD96 pKa = 4.71VDD98 pKa = 5.98DD99 pKa = 5.83IPDD102 pKa = 5.13DD103 pKa = 4.53IDD105 pKa = 6.11DD106 pKa = 4.57EE107 pKa = 4.8DD108 pKa = 4.95VNDD111 pKa = 5.07DD112 pKa = 3.93GNINTSSIGNQIRR125 pKa = 11.84CIVIHH130 pKa = 6.3NNPGPHH136 pKa = 6.25MSFIDD141 pKa = 3.47PDD143 pKa = 3.47AAYY146 pKa = 10.42VAEE149 pKa = 4.31IPKK152 pKa = 10.45
MM1 pKa = 7.51IALYY5 pKa = 10.6CEE7 pKa = 4.05NRR9 pKa = 11.84SDD11 pKa = 3.51KK12 pKa = 10.65YY13 pKa = 11.56APIHH17 pKa = 6.41LFAEE21 pKa = 4.49LAGMKK26 pKa = 10.12QNEE29 pKa = 4.59DD30 pKa = 2.96FTAYY34 pKa = 10.38GEE36 pKa = 3.99EE37 pKa = 4.01HH38 pKa = 6.97RR39 pKa = 11.84AQEE42 pKa = 4.68LCMMAPISYY51 pKa = 10.13VDD53 pKa = 3.44SEE55 pKa = 4.25LTIRR59 pKa = 11.84GINIDD64 pKa = 4.25LNVTPDD70 pKa = 3.18IDD72 pKa = 4.01VVGDD76 pKa = 3.81DD77 pKa = 5.53GYY79 pKa = 11.52DD80 pKa = 3.37SSDD83 pKa = 3.43PCDD86 pKa = 3.33QKK88 pKa = 11.51VDD90 pKa = 3.71SDD92 pKa = 3.86NDD94 pKa = 3.82LDD96 pKa = 4.71VDD98 pKa = 5.98DD99 pKa = 5.83IPDD102 pKa = 5.13DD103 pKa = 4.53IDD105 pKa = 6.11DD106 pKa = 4.57EE107 pKa = 4.8DD108 pKa = 4.95VNDD111 pKa = 5.07DD112 pKa = 3.93GNINTSSIGNQIRR125 pKa = 11.84CIVIHH130 pKa = 6.3NNPGPHH136 pKa = 6.25MSFIDD141 pKa = 3.47PDD143 pKa = 3.47AAYY146 pKa = 10.42VAEE149 pKa = 4.31IPKK152 pKa = 10.45
Molecular weight: 16.84 kDa
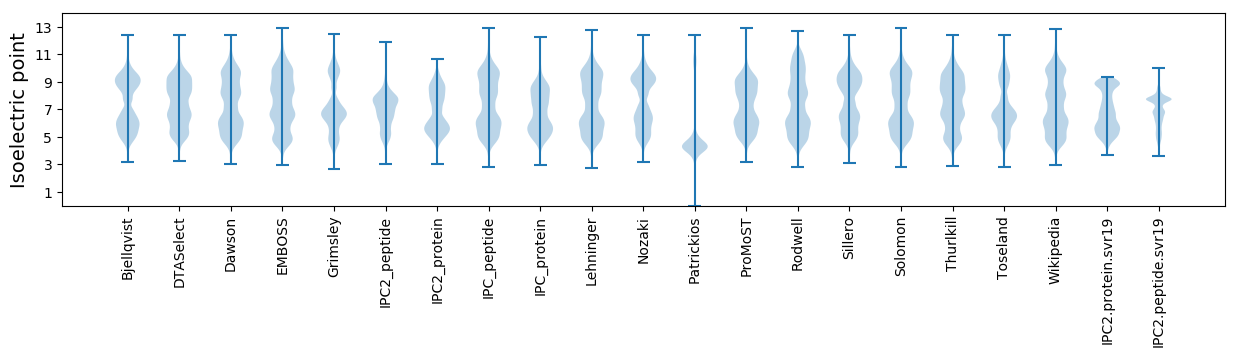
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A2P5YPA9|A0A2P5YPA9_GOSBA F-box domain-containing protein OS=Gossypium barbadense OX=3634 GN=ES319_A01G165000v1 PE=4 SV=1
MM1 pKa = 7.08VSKK4 pKa = 10.96APAHH8 pKa = 6.14TSTKK12 pKa = 10.51VPMLLEE18 pKa = 4.57APAHH22 pKa = 6.11PRR24 pKa = 11.84HH25 pKa = 5.9QGADD29 pKa = 3.35GAQGSRR35 pKa = 11.84TTKK38 pKa = 10.01VADD41 pKa = 3.69GARR44 pKa = 11.84GSRR47 pKa = 11.84TTRR50 pKa = 11.84GLGCRR55 pKa = 11.84WCPRR59 pKa = 11.84LPHH62 pKa = 6.5DD63 pKa = 4.1QGPRR67 pKa = 11.84LPMVSEE73 pKa = 4.26APAYY77 pKa = 8.86PSHH80 pKa = 6.49QGADD84 pKa = 3.46AARR87 pKa = 11.84GSCTTNVLGVQVVRR101 pKa = 11.84PRR103 pKa = 11.84LQHH106 pKa = 4.89VQAAMGGCFGTIDD119 pKa = 3.31VLGYY123 pKa = 8.0QWCLMLRR130 pKa = 11.84RR131 pKa = 11.84DD132 pKa = 3.93QGPKK136 pKa = 9.44VPEE139 pKa = 3.75VLVPKK144 pKa = 10.84GEE146 pKa = 4.17GPGWFEE152 pKa = 4.57KK153 pKa = 10.93GGTGTTCSIIFIFQNGSPEE172 pKa = 3.76HH173 pKa = 6.25RR174 pKa = 11.84RR175 pKa = 11.84EE176 pKa = 3.89SSLDD180 pKa = 3.06HH181 pKa = 6.59HH182 pKa = 7.33RR183 pKa = 11.84NGSLATFMCLKK194 pKa = 9.87QGYY197 pKa = 9.59FMVPTLLHH205 pKa = 5.15QRR207 pKa = 11.84EE208 pKa = 4.26PKK210 pKa = 9.95ISFDD214 pKa = 3.42ARR216 pKa = 11.84PSRR219 pKa = 11.84TSDD222 pKa = 2.98APRR225 pKa = 11.84CRR227 pKa = 11.84CCPMLRR233 pKa = 11.84HH234 pKa = 6.05IKK236 pKa = 10.14GAGGARR242 pKa = 11.84WPCTTGWHH250 pKa = 5.78TLPIVYY256 pKa = 8.73EE257 pKa = 3.98ATARR261 pKa = 11.84PGAKK265 pKa = 9.81GAGGSCCKK273 pKa = 10.47RR274 pKa = 11.84GGTWMIHH281 pKa = 5.17SLLGTGHH288 pKa = 6.01ATIFEE293 pKa = 4.51SYY295 pKa = 10.33LWDD298 pKa = 3.94DD299 pKa = 3.66SFIIGDD305 pKa = 4.35RR306 pKa = 11.84FQMSEE311 pKa = 3.34IGTLFSAQRR320 pKa = 11.84SCTSEE325 pKa = 3.88APRR328 pKa = 11.84CRR330 pKa = 11.84CCSMTRR336 pKa = 11.84TMGWPPLRR344 pKa = 11.84ILPQTPKK351 pKa = 9.93IEE353 pKa = 4.23QVPEE357 pKa = 4.13PPTVEE362 pKa = 4.22SSHH365 pKa = 5.76SHH367 pKa = 5.02HH368 pKa = 7.12HH369 pKa = 6.77DD370 pKa = 2.93PRR372 pKa = 11.84YY373 pKa = 9.02ATVSRR378 pKa = 11.84GGGTANKK385 pKa = 8.31ATLPLTIPRR394 pKa = 11.84RR395 pKa = 11.84VFKK398 pKa = 11.0SSAKK402 pKa = 10.27DD403 pKa = 3.36FTHH406 pKa = 6.74RR407 pKa = 11.84SVGITLQGGTRR418 pKa = 11.84DD419 pKa = 3.52LSAAIAAPMTRR430 pKa = 11.84AIGGQRR436 pKa = 11.84PLMRR440 pKa = 11.84VGNWATGPCIASIPDD455 pKa = 3.28SDD457 pKa = 5.6LEE459 pKa = 4.21AFSYY463 pKa = 11.0NPAHH467 pKa = 6.37GRR469 pKa = 11.84ADD471 pKa = 3.39IEE473 pKa = 4.46GSKK476 pKa = 11.16SNVAMNAWLPQASYY490 pKa = 10.49PCGNFSDD497 pKa = 3.71TSSFKK502 pKa = 10.28FRR504 pKa = 11.84RR505 pKa = 11.84SKK507 pKa = 11.22GSIGHH512 pKa = 7.39AFTPNASAPTGAEE525 pKa = 3.98VVDD528 pKa = 4.27GARR531 pKa = 11.84GSRR534 pKa = 11.84TSEE537 pKa = 3.98APSKK541 pKa = 10.18ATLPLTIPRR550 pKa = 11.84RR551 pKa = 11.84VFKK554 pKa = 10.99SSAKK558 pKa = 10.07DD559 pKa = 3.14STRR562 pKa = 11.84RR563 pKa = 11.84SVGITLQGGARR574 pKa = 11.84DD575 pKa = 4.05LSAAAAAPTTRR586 pKa = 11.84ALGGQRR592 pKa = 11.84PLLRR596 pKa = 11.84VGNRR600 pKa = 11.84ATGACVASSPDD611 pKa = 3.19SDD613 pKa = 5.32LEE615 pKa = 4.35AFSHH619 pKa = 6.03NPAHH623 pKa = 7.32GSFAPLAFQPSAMTNCANQRR643 pKa = 11.84SEE645 pKa = 4.2STVRR649 pKa = 11.84RR650 pKa = 11.84PGKK653 pKa = 10.24APEE656 pKa = 4.17GVIPSPSPDD665 pKa = 3.04QHH667 pKa = 7.47AVTCSRR673 pKa = 11.84RR674 pKa = 11.84GSSSSSAPVADD685 pKa = 4.06EE686 pKa = 4.75FGTGTPVRR694 pKa = 11.84SPQSQSFSRR703 pKa = 11.84GYY705 pKa = 11.09GSIFPTSLAYY715 pKa = 9.23IVPLTRR721 pKa = 11.84GCSPWRR727 pKa = 11.84PDD729 pKa = 2.8AVMNISTATVMARR742 pKa = 11.84GRR744 pKa = 11.84CGSFSRR750 pKa = 11.84DD751 pKa = 2.64TRR753 pKa = 11.84QWLRR757 pKa = 11.84RR758 pKa = 11.84GTYY761 pKa = 8.76GGHH764 pKa = 6.1RR765 pKa = 11.84KK766 pKa = 8.93VAGAGKK772 pKa = 10.33RR773 pKa = 11.84LFRR776 pKa = 11.84PLGVQNRR783 pKa = 11.84APLRR787 pKa = 11.84CLRR790 pKa = 11.84RR791 pKa = 11.84KK792 pKa = 9.95APRR795 pKa = 11.84LRR797 pKa = 11.84VPKK800 pKa = 10.11RR801 pKa = 11.84LASSRR806 pKa = 11.84GSLTFSPLFPPQLNVLVSSSRR827 pKa = 11.84SAFQPEE833 pKa = 4.44SLGCRR838 pKa = 11.84YY839 pKa = 9.66GHH841 pKa = 7.22LPAAIDD847 pKa = 3.32SGAEE851 pKa = 3.53KK852 pKa = 10.34RR853 pKa = 11.84GPGRR857 pKa = 11.84EE858 pKa = 3.81ASRR861 pKa = 11.84SSRR864 pKa = 11.84SSNAQSGVISSHH876 pKa = 6.0ALYY879 pKa = 9.5PSHH882 pKa = 7.04AALPPSVKK890 pKa = 10.1PSGSAPAINAASRR903 pKa = 11.84TSLKK907 pKa = 10.3GSCSLTPGGAASPAGRR923 pKa = 11.84EE924 pKa = 4.2HH925 pKa = 6.86TSQLAAGCRR934 pKa = 11.84PMLLVVTIRR943 pKa = 11.84CSTGLDD949 pKa = 2.88LGLCHH954 pKa = 7.14FEE956 pKa = 3.9IHH958 pKa = 6.51HH959 pKa = 6.22SAGSCTCPLKK969 pKa = 10.13TPRR972 pKa = 11.84KK973 pKa = 7.32RR974 pKa = 11.84SRR976 pKa = 11.84RR977 pKa = 11.84IRR979 pKa = 11.84NN980 pKa = 3.17
MM1 pKa = 7.08VSKK4 pKa = 10.96APAHH8 pKa = 6.14TSTKK12 pKa = 10.51VPMLLEE18 pKa = 4.57APAHH22 pKa = 6.11PRR24 pKa = 11.84HH25 pKa = 5.9QGADD29 pKa = 3.35GAQGSRR35 pKa = 11.84TTKK38 pKa = 10.01VADD41 pKa = 3.69GARR44 pKa = 11.84GSRR47 pKa = 11.84TTRR50 pKa = 11.84GLGCRR55 pKa = 11.84WCPRR59 pKa = 11.84LPHH62 pKa = 6.5DD63 pKa = 4.1QGPRR67 pKa = 11.84LPMVSEE73 pKa = 4.26APAYY77 pKa = 8.86PSHH80 pKa = 6.49QGADD84 pKa = 3.46AARR87 pKa = 11.84GSCTTNVLGVQVVRR101 pKa = 11.84PRR103 pKa = 11.84LQHH106 pKa = 4.89VQAAMGGCFGTIDD119 pKa = 3.31VLGYY123 pKa = 8.0QWCLMLRR130 pKa = 11.84RR131 pKa = 11.84DD132 pKa = 3.93QGPKK136 pKa = 9.44VPEE139 pKa = 3.75VLVPKK144 pKa = 10.84GEE146 pKa = 4.17GPGWFEE152 pKa = 4.57KK153 pKa = 10.93GGTGTTCSIIFIFQNGSPEE172 pKa = 3.76HH173 pKa = 6.25RR174 pKa = 11.84RR175 pKa = 11.84EE176 pKa = 3.89SSLDD180 pKa = 3.06HH181 pKa = 6.59HH182 pKa = 7.33RR183 pKa = 11.84NGSLATFMCLKK194 pKa = 9.87QGYY197 pKa = 9.59FMVPTLLHH205 pKa = 5.15QRR207 pKa = 11.84EE208 pKa = 4.26PKK210 pKa = 9.95ISFDD214 pKa = 3.42ARR216 pKa = 11.84PSRR219 pKa = 11.84TSDD222 pKa = 2.98APRR225 pKa = 11.84CRR227 pKa = 11.84CCPMLRR233 pKa = 11.84HH234 pKa = 6.05IKK236 pKa = 10.14GAGGARR242 pKa = 11.84WPCTTGWHH250 pKa = 5.78TLPIVYY256 pKa = 8.73EE257 pKa = 3.98ATARR261 pKa = 11.84PGAKK265 pKa = 9.81GAGGSCCKK273 pKa = 10.47RR274 pKa = 11.84GGTWMIHH281 pKa = 5.17SLLGTGHH288 pKa = 6.01ATIFEE293 pKa = 4.51SYY295 pKa = 10.33LWDD298 pKa = 3.94DD299 pKa = 3.66SFIIGDD305 pKa = 4.35RR306 pKa = 11.84FQMSEE311 pKa = 3.34IGTLFSAQRR320 pKa = 11.84SCTSEE325 pKa = 3.88APRR328 pKa = 11.84CRR330 pKa = 11.84CCSMTRR336 pKa = 11.84TMGWPPLRR344 pKa = 11.84ILPQTPKK351 pKa = 9.93IEE353 pKa = 4.23QVPEE357 pKa = 4.13PPTVEE362 pKa = 4.22SSHH365 pKa = 5.76SHH367 pKa = 5.02HH368 pKa = 7.12HH369 pKa = 6.77DD370 pKa = 2.93PRR372 pKa = 11.84YY373 pKa = 9.02ATVSRR378 pKa = 11.84GGGTANKK385 pKa = 8.31ATLPLTIPRR394 pKa = 11.84RR395 pKa = 11.84VFKK398 pKa = 11.0SSAKK402 pKa = 10.27DD403 pKa = 3.36FTHH406 pKa = 6.74RR407 pKa = 11.84SVGITLQGGTRR418 pKa = 11.84DD419 pKa = 3.52LSAAIAAPMTRR430 pKa = 11.84AIGGQRR436 pKa = 11.84PLMRR440 pKa = 11.84VGNWATGPCIASIPDD455 pKa = 3.28SDD457 pKa = 5.6LEE459 pKa = 4.21AFSYY463 pKa = 11.0NPAHH467 pKa = 6.37GRR469 pKa = 11.84ADD471 pKa = 3.39IEE473 pKa = 4.46GSKK476 pKa = 11.16SNVAMNAWLPQASYY490 pKa = 10.49PCGNFSDD497 pKa = 3.71TSSFKK502 pKa = 10.28FRR504 pKa = 11.84RR505 pKa = 11.84SKK507 pKa = 11.22GSIGHH512 pKa = 7.39AFTPNASAPTGAEE525 pKa = 3.98VVDD528 pKa = 4.27GARR531 pKa = 11.84GSRR534 pKa = 11.84TSEE537 pKa = 3.98APSKK541 pKa = 10.18ATLPLTIPRR550 pKa = 11.84RR551 pKa = 11.84VFKK554 pKa = 10.99SSAKK558 pKa = 10.07DD559 pKa = 3.14STRR562 pKa = 11.84RR563 pKa = 11.84SVGITLQGGARR574 pKa = 11.84DD575 pKa = 4.05LSAAAAAPTTRR586 pKa = 11.84ALGGQRR592 pKa = 11.84PLLRR596 pKa = 11.84VGNRR600 pKa = 11.84ATGACVASSPDD611 pKa = 3.19SDD613 pKa = 5.32LEE615 pKa = 4.35AFSHH619 pKa = 6.03NPAHH623 pKa = 7.32GSFAPLAFQPSAMTNCANQRR643 pKa = 11.84SEE645 pKa = 4.2STVRR649 pKa = 11.84RR650 pKa = 11.84PGKK653 pKa = 10.24APEE656 pKa = 4.17GVIPSPSPDD665 pKa = 3.04QHH667 pKa = 7.47AVTCSRR673 pKa = 11.84RR674 pKa = 11.84GSSSSSAPVADD685 pKa = 4.06EE686 pKa = 4.75FGTGTPVRR694 pKa = 11.84SPQSQSFSRR703 pKa = 11.84GYY705 pKa = 11.09GSIFPTSLAYY715 pKa = 9.23IVPLTRR721 pKa = 11.84GCSPWRR727 pKa = 11.84PDD729 pKa = 2.8AVMNISTATVMARR742 pKa = 11.84GRR744 pKa = 11.84CGSFSRR750 pKa = 11.84DD751 pKa = 2.64TRR753 pKa = 11.84QWLRR757 pKa = 11.84RR758 pKa = 11.84GTYY761 pKa = 8.76GGHH764 pKa = 6.1RR765 pKa = 11.84KK766 pKa = 8.93VAGAGKK772 pKa = 10.33RR773 pKa = 11.84LFRR776 pKa = 11.84PLGVQNRR783 pKa = 11.84APLRR787 pKa = 11.84CLRR790 pKa = 11.84RR791 pKa = 11.84KK792 pKa = 9.95APRR795 pKa = 11.84LRR797 pKa = 11.84VPKK800 pKa = 10.11RR801 pKa = 11.84LASSRR806 pKa = 11.84GSLTFSPLFPPQLNVLVSSSRR827 pKa = 11.84SAFQPEE833 pKa = 4.44SLGCRR838 pKa = 11.84YY839 pKa = 9.66GHH841 pKa = 7.22LPAAIDD847 pKa = 3.32SGAEE851 pKa = 3.53KK852 pKa = 10.34RR853 pKa = 11.84GPGRR857 pKa = 11.84EE858 pKa = 3.81ASRR861 pKa = 11.84SSRR864 pKa = 11.84SSNAQSGVISSHH876 pKa = 6.0ALYY879 pKa = 9.5PSHH882 pKa = 7.04AALPPSVKK890 pKa = 10.1PSGSAPAINAASRR903 pKa = 11.84TSLKK907 pKa = 10.3GSCSLTPGGAASPAGRR923 pKa = 11.84EE924 pKa = 4.2HH925 pKa = 6.86TSQLAAGCRR934 pKa = 11.84PMLLVVTIRR943 pKa = 11.84CSTGLDD949 pKa = 2.88LGLCHH954 pKa = 7.14FEE956 pKa = 3.9IHH958 pKa = 6.51HH959 pKa = 6.22SAGSCTCPLKK969 pKa = 10.13TPRR972 pKa = 11.84KK973 pKa = 7.32RR974 pKa = 11.84SRR976 pKa = 11.84RR977 pKa = 11.84IRR979 pKa = 11.84NN980 pKa = 3.17
Molecular weight: 104.46 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
15047206 |
26 |
6014 |
375.7 |
41.98 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.547 ± 0.011 | 1.91 ± 0.006 |
5.195 ± 0.008 | 6.437 ± 0.016 |
4.244 ± 0.009 | 6.503 ± 0.011 |
2.407 ± 0.005 | 5.413 ± 0.009 |
6.229 ± 0.012 | 9.773 ± 0.015 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.494 ± 0.005 | 4.634 ± 0.009 |
4.938 ± 0.012 | 3.694 ± 0.008 |
5.245 ± 0.01 | 8.8 ± 0.015 |
4.957 ± 0.008 | 6.479 ± 0.009 |
1.315 ± 0.004 | 2.774 ± 0.007 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
