
Lactobacillus prophage Lj965
Taxonomy: Viruses; Duplodnaviria; Heunggongvirae; Uroviricota; Caudoviricetes; Caudovirales; Siphoviridae; unclassified Siphoviridae
Average proteome isoelectric point is 6.75
Get precalculated fractions of proteins
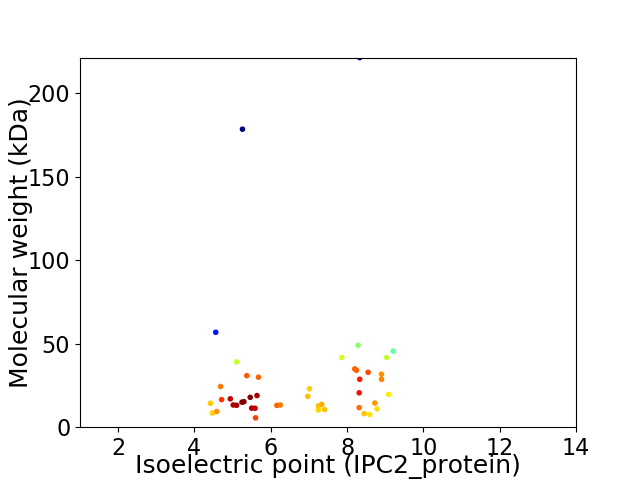
Virtual 2D-PAGE plot for 46 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions


Protein with the lowest isoelectric point:
>tr|Q6SE96|Q6SE96_9CAUD Uncharacterized protein OS=Lactobacillus prophage Lj965 OX=139870 GN=Ljo_0296b PE=4 SV=1
MM1 pKa = 7.77KK2 pKa = 10.4EE3 pKa = 3.69FDD5 pKa = 5.68PRR7 pKa = 11.84DD8 pKa = 3.25LWKK11 pKa = 10.42LQEE14 pKa = 4.14VNGMVLRR21 pKa = 11.84DD22 pKa = 3.16IHH24 pKa = 7.53GIDD27 pKa = 3.38VAIGKK32 pKa = 9.63GFEE35 pKa = 4.16YY36 pKa = 10.89KK37 pKa = 10.42NIKK40 pKa = 10.31AFIEE44 pKa = 4.81VYY46 pKa = 6.45TTEE49 pKa = 4.24YY50 pKa = 10.34GVKK53 pKa = 10.56DD54 pKa = 4.08FMEE57 pKa = 4.38KK58 pKa = 9.55MGFEE62 pKa = 3.99NSEE65 pKa = 4.54DD66 pKa = 3.29FTKK69 pKa = 10.98YY70 pKa = 10.23YY71 pKa = 10.66FKK73 pKa = 10.76EE74 pKa = 4.45FPDD77 pKa = 3.43EE78 pKa = 4.29CDD80 pKa = 3.52WYY82 pKa = 10.41DD83 pKa = 3.1ACYY86 pKa = 9.32WAFNGIYY93 pKa = 10.3ADD95 pKa = 4.31DD96 pKa = 4.53LALKK100 pKa = 10.24GYY102 pKa = 9.15EE103 pKa = 3.87EE104 pKa = 4.17EE105 pKa = 5.07AYY107 pKa = 10.98LDD109 pKa = 4.07AEE111 pKa = 4.22DD112 pKa = 4.44AKK114 pKa = 10.79RR115 pKa = 11.84DD116 pKa = 3.81RR117 pKa = 11.84LAGKK121 pKa = 9.73
MM1 pKa = 7.77KK2 pKa = 10.4EE3 pKa = 3.69FDD5 pKa = 5.68PRR7 pKa = 11.84DD8 pKa = 3.25LWKK11 pKa = 10.42LQEE14 pKa = 4.14VNGMVLRR21 pKa = 11.84DD22 pKa = 3.16IHH24 pKa = 7.53GIDD27 pKa = 3.38VAIGKK32 pKa = 9.63GFEE35 pKa = 4.16YY36 pKa = 10.89KK37 pKa = 10.42NIKK40 pKa = 10.31AFIEE44 pKa = 4.81VYY46 pKa = 6.45TTEE49 pKa = 4.24YY50 pKa = 10.34GVKK53 pKa = 10.56DD54 pKa = 4.08FMEE57 pKa = 4.38KK58 pKa = 9.55MGFEE62 pKa = 3.99NSEE65 pKa = 4.54DD66 pKa = 3.29FTKK69 pKa = 10.98YY70 pKa = 10.23YY71 pKa = 10.66FKK73 pKa = 10.76EE74 pKa = 4.45FPDD77 pKa = 3.43EE78 pKa = 4.29CDD80 pKa = 3.52WYY82 pKa = 10.41DD83 pKa = 3.1ACYY86 pKa = 9.32WAFNGIYY93 pKa = 10.3ADD95 pKa = 4.31DD96 pKa = 4.53LALKK100 pKa = 10.24GYY102 pKa = 9.15EE103 pKa = 3.87EE104 pKa = 4.17EE105 pKa = 5.07AYY107 pKa = 10.98LDD109 pKa = 4.07AEE111 pKa = 4.22DD112 pKa = 4.44AKK114 pKa = 10.79RR115 pKa = 11.84DD116 pKa = 3.81RR117 pKa = 11.84LAGKK121 pKa = 9.73
Molecular weight: 14.34 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|Q6SE82|Q6SE82_9CAUD Uncharacterized protein OS=Lactobacillus prophage Lj965 OX=139870 GN=Ljo_0310 PE=4 SV=1
MM1 pKa = 7.46KK2 pKa = 10.42VNKK5 pKa = 10.08KK6 pKa = 10.33KK7 pKa = 9.25FTYY10 pKa = 8.48WQLRR14 pKa = 11.84DD15 pKa = 3.71LQDD18 pKa = 3.23EE19 pKa = 4.14QRR21 pKa = 11.84NQDD24 pKa = 3.34EE25 pKa = 4.13ATEE28 pKa = 3.94RR29 pKa = 11.84LKK31 pKa = 10.72IINNAYY37 pKa = 10.12QKK39 pKa = 10.14AQSYY43 pKa = 10.86LSDD46 pKa = 3.85EE47 pKa = 4.07VQKK50 pKa = 10.48IYY52 pKa = 10.87RR53 pKa = 11.84RR54 pKa = 11.84YY55 pKa = 9.46FYY57 pKa = 11.46ADD59 pKa = 2.82ISKK62 pKa = 11.19DD63 pKa = 3.42EE64 pKa = 4.26VATIMSSHH72 pKa = 7.08ISPSEE77 pKa = 3.77LVTLKK82 pKa = 10.78ALSSSITDD90 pKa = 3.4KK91 pKa = 10.93EE92 pKa = 4.08SRR94 pKa = 11.84QAVDD98 pKa = 3.25NYY100 pKa = 10.8LSRR103 pKa = 11.84LAAKK107 pKa = 10.3SRR109 pKa = 11.84ITRR112 pKa = 11.84LEE114 pKa = 3.68EE115 pKa = 3.91MQLKK119 pKa = 10.42AYY121 pKa = 9.57IAAKK125 pKa = 10.08SAGATEE131 pKa = 4.8LDD133 pKa = 3.92QNVKK137 pKa = 10.15LHH139 pKa = 5.8TDD141 pKa = 2.49IMKK144 pKa = 10.36RR145 pKa = 11.84AWSEE149 pKa = 3.99AEE151 pKa = 4.12KK152 pKa = 10.79QSAVYY157 pKa = 8.24DD158 pKa = 3.7TTKK161 pKa = 10.84DD162 pKa = 3.64YY163 pKa = 10.28TLHH166 pKa = 6.46SPHH169 pKa = 7.08SVEE172 pKa = 4.17VKK174 pKa = 9.58QDD176 pKa = 3.35KK177 pKa = 10.57IVIKK181 pKa = 11.0NPDD184 pKa = 3.33TGKK187 pKa = 10.49EE188 pKa = 4.14VATVPMDD195 pKa = 3.62KK196 pKa = 10.52DD197 pKa = 3.86VPKK200 pKa = 10.99SKK202 pKa = 8.96ITEE205 pKa = 3.76IPNRR209 pKa = 11.84YY210 pKa = 8.47VEE212 pKa = 4.54KK213 pKa = 10.73SLEE216 pKa = 4.21TRR218 pKa = 11.84WKK220 pKa = 10.16GKK222 pKa = 9.96NFSARR227 pKa = 11.84IWGNTDD233 pKa = 2.67KK234 pKa = 11.03LAEE237 pKa = 4.17RR238 pKa = 11.84LQEE241 pKa = 4.02LFTVKK246 pKa = 10.08EE247 pKa = 3.97LSNLPEE253 pKa = 4.95RR254 pKa = 11.84EE255 pKa = 3.85MIKK258 pKa = 10.33RR259 pKa = 11.84IEE261 pKa = 3.84QEE263 pKa = 3.92FNVGKK268 pKa = 10.02FYY270 pKa = 11.07ASRR273 pKa = 11.84LIRR276 pKa = 11.84TEE278 pKa = 4.25ANFFYY283 pKa = 11.13SKK285 pKa = 10.52IKK287 pKa = 10.06LDD289 pKa = 3.25NWRR292 pKa = 11.84KK293 pKa = 10.06RR294 pKa = 11.84GVKK297 pKa = 9.51QYY299 pKa = 11.32QLLAVIDD306 pKa = 3.92SRR308 pKa = 11.84TSKK311 pKa = 9.87ICRR314 pKa = 11.84SINGNIYY321 pKa = 9.51NVKK324 pKa = 10.43DD325 pKa = 3.37AVFGKK330 pKa = 9.85NVPPLHH336 pKa = 6.12PFCRR340 pKa = 11.84TVPVIYY346 pKa = 10.43LGNARR351 pKa = 11.84SANNKK356 pKa = 8.44PVKK359 pKa = 10.15KK360 pKa = 10.59
MM1 pKa = 7.46KK2 pKa = 10.42VNKK5 pKa = 10.08KK6 pKa = 10.33KK7 pKa = 9.25FTYY10 pKa = 8.48WQLRR14 pKa = 11.84DD15 pKa = 3.71LQDD18 pKa = 3.23EE19 pKa = 4.14QRR21 pKa = 11.84NQDD24 pKa = 3.34EE25 pKa = 4.13ATEE28 pKa = 3.94RR29 pKa = 11.84LKK31 pKa = 10.72IINNAYY37 pKa = 10.12QKK39 pKa = 10.14AQSYY43 pKa = 10.86LSDD46 pKa = 3.85EE47 pKa = 4.07VQKK50 pKa = 10.48IYY52 pKa = 10.87RR53 pKa = 11.84RR54 pKa = 11.84YY55 pKa = 9.46FYY57 pKa = 11.46ADD59 pKa = 2.82ISKK62 pKa = 11.19DD63 pKa = 3.42EE64 pKa = 4.26VATIMSSHH72 pKa = 7.08ISPSEE77 pKa = 3.77LVTLKK82 pKa = 10.78ALSSSITDD90 pKa = 3.4KK91 pKa = 10.93EE92 pKa = 4.08SRR94 pKa = 11.84QAVDD98 pKa = 3.25NYY100 pKa = 10.8LSRR103 pKa = 11.84LAAKK107 pKa = 10.3SRR109 pKa = 11.84ITRR112 pKa = 11.84LEE114 pKa = 3.68EE115 pKa = 3.91MQLKK119 pKa = 10.42AYY121 pKa = 9.57IAAKK125 pKa = 10.08SAGATEE131 pKa = 4.8LDD133 pKa = 3.92QNVKK137 pKa = 10.15LHH139 pKa = 5.8TDD141 pKa = 2.49IMKK144 pKa = 10.36RR145 pKa = 11.84AWSEE149 pKa = 3.99AEE151 pKa = 4.12KK152 pKa = 10.79QSAVYY157 pKa = 8.24DD158 pKa = 3.7TTKK161 pKa = 10.84DD162 pKa = 3.64YY163 pKa = 10.28TLHH166 pKa = 6.46SPHH169 pKa = 7.08SVEE172 pKa = 4.17VKK174 pKa = 9.58QDD176 pKa = 3.35KK177 pKa = 10.57IVIKK181 pKa = 11.0NPDD184 pKa = 3.33TGKK187 pKa = 10.49EE188 pKa = 4.14VATVPMDD195 pKa = 3.62KK196 pKa = 10.52DD197 pKa = 3.86VPKK200 pKa = 10.99SKK202 pKa = 8.96ITEE205 pKa = 3.76IPNRR209 pKa = 11.84YY210 pKa = 8.47VEE212 pKa = 4.54KK213 pKa = 10.73SLEE216 pKa = 4.21TRR218 pKa = 11.84WKK220 pKa = 10.16GKK222 pKa = 9.96NFSARR227 pKa = 11.84IWGNTDD233 pKa = 2.67KK234 pKa = 11.03LAEE237 pKa = 4.17RR238 pKa = 11.84LQEE241 pKa = 4.02LFTVKK246 pKa = 10.08EE247 pKa = 3.97LSNLPEE253 pKa = 4.95RR254 pKa = 11.84EE255 pKa = 3.85MIKK258 pKa = 10.33RR259 pKa = 11.84IEE261 pKa = 3.84QEE263 pKa = 3.92FNVGKK268 pKa = 10.02FYY270 pKa = 11.07ASRR273 pKa = 11.84LIRR276 pKa = 11.84TEE278 pKa = 4.25ANFFYY283 pKa = 11.13SKK285 pKa = 10.52IKK287 pKa = 10.06LDD289 pKa = 3.25NWRR292 pKa = 11.84KK293 pKa = 10.06RR294 pKa = 11.84GVKK297 pKa = 9.51QYY299 pKa = 11.32QLLAVIDD306 pKa = 3.92SRR308 pKa = 11.84TSKK311 pKa = 9.87ICRR314 pKa = 11.84SINGNIYY321 pKa = 9.51NVKK324 pKa = 10.43DD325 pKa = 3.37AVFGKK330 pKa = 9.85NVPPLHH336 pKa = 6.12PFCRR340 pKa = 11.84TVPVIYY346 pKa = 10.43LGNARR351 pKa = 11.84SANNKK356 pKa = 8.44PVKK359 pKa = 10.15KK360 pKa = 10.59
Molecular weight: 41.71 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
11916 |
49 |
2021 |
259.0 |
29.27 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.276 ± 0.474 | 0.529 ± 0.126 |
6.596 ± 0.369 | 6.185 ± 0.389 |
4.087 ± 0.27 | 6.16 ± 0.685 |
1.435 ± 0.172 | 6.63 ± 0.225 |
8.87 ± 0.568 | 8.124 ± 0.318 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.266 ± 0.216 | 6.781 ± 0.435 |
2.87 ± 0.212 | 4.397 ± 0.232 |
4.045 ± 0.217 | 6.73 ± 0.359 |
5.975 ± 0.426 | 5.715 ± 0.228 |
1.502 ± 0.146 | 3.827 ± 0.376 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
