
Ctenophore-associated circular virus 2
Taxonomy: Viruses; unclassified viruses; unclassified DNA viruses; unclassified ssDNA viruses
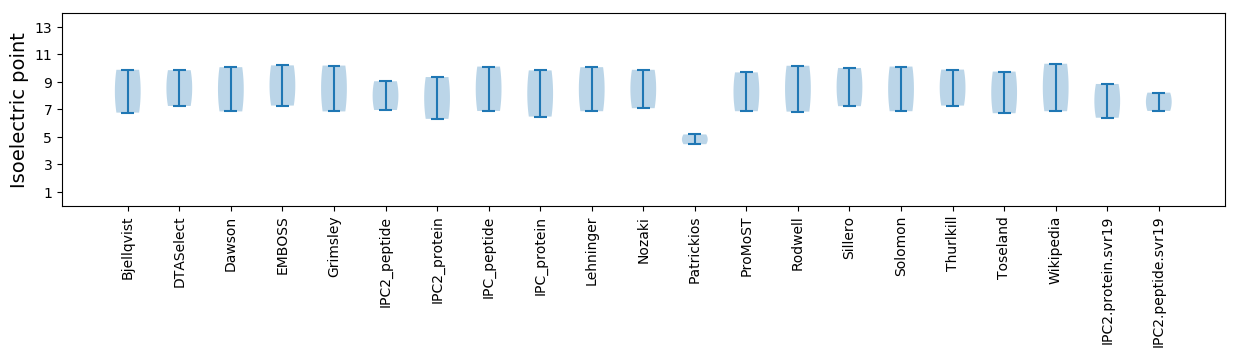
Average proteome isoelectric point is 7.62
Get precalculated fractions of proteins
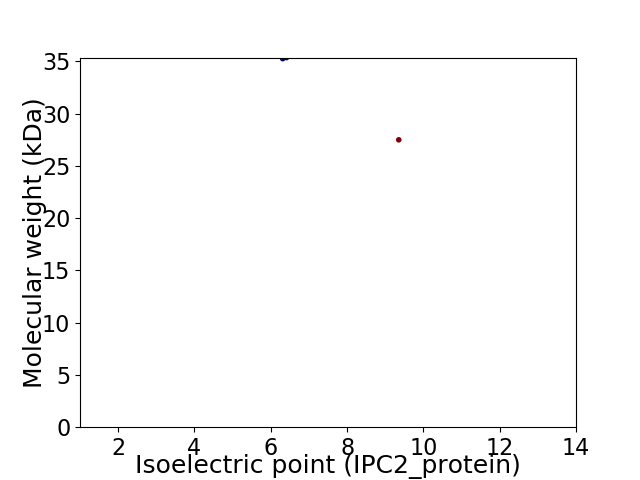
Virtual 2D-PAGE plot for 2 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A141MJA8|A0A141MJA8_9VIRU Uncharacterized protein OS=Ctenophore-associated circular virus 2 OX=1778559 PE=4 SV=1
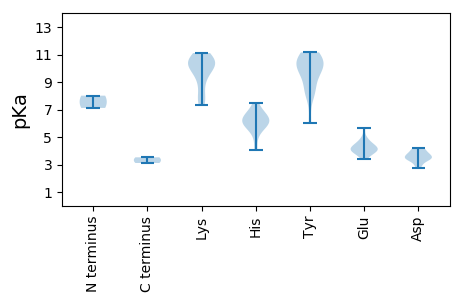
MM1 pKa = 8.01PSPKK5 pKa = 9.1STRR8 pKa = 11.84WCFTHH13 pKa = 6.62NNWTEE18 pKa = 3.71PVFQQWSNLLGDD30 pKa = 3.7EE31 pKa = 4.59ANVKK35 pKa = 10.37FGIIGKK41 pKa = 8.85EE42 pKa = 3.9VGEE45 pKa = 4.24QGTPHH50 pKa = 6.3LQGFLILHH58 pKa = 5.86RR59 pKa = 11.84QQRR62 pKa = 11.84LSWVRR67 pKa = 11.84RR68 pKa = 11.84HH69 pKa = 6.21FPDD72 pKa = 4.16GCHH75 pKa = 5.59WSIARR80 pKa = 11.84SDD82 pKa = 3.77SEE84 pKa = 4.64TNRR87 pKa = 11.84TYY89 pKa = 10.8CKK91 pKa = 10.14KK92 pKa = 10.66DD93 pKa = 2.75GDD95 pKa = 4.2FIEE98 pKa = 5.64FGVFPDD104 pKa = 3.83AQGKK108 pKa = 7.76RR109 pKa = 11.84TDD111 pKa = 3.61LDD113 pKa = 4.1QFIEE117 pKa = 4.01WLDD120 pKa = 3.59EE121 pKa = 4.24FEE123 pKa = 4.99SNNGRR128 pKa = 11.84PASSPEE134 pKa = 3.53IAKK137 pKa = 7.36THH139 pKa = 5.86PKK141 pKa = 9.64MYY143 pKa = 10.01LRR145 pKa = 11.84YY146 pKa = 8.26PRR148 pKa = 11.84SVRR151 pKa = 11.84LAKK154 pKa = 10.09RR155 pKa = 11.84RR156 pKa = 11.84CALFPVQEE164 pKa = 4.52GDD166 pKa = 3.52LNDD169 pKa = 3.53WQRR172 pKa = 11.84EE173 pKa = 4.19LEE175 pKa = 4.37EE176 pKa = 4.85KK177 pKa = 10.8LGADD181 pKa = 3.34PDD183 pKa = 3.95DD184 pKa = 3.79RR185 pKa = 11.84TVLFYY190 pKa = 11.16VDD192 pKa = 3.62PDD194 pKa = 3.57GGKK197 pKa = 10.12GKK199 pKa = 7.61TWFVRR204 pKa = 11.84RR205 pKa = 11.84YY206 pKa = 8.26LTLHH210 pKa = 6.77PNLAQALPIGQIKK223 pKa = 10.55DD224 pKa = 3.03MAYY227 pKa = 10.29AVDD230 pKa = 3.42TNARR234 pKa = 11.84VFFVNVARR242 pKa = 11.84SGMEE246 pKa = 3.38FLPYY250 pKa = 10.26RR251 pKa = 11.84FLEE254 pKa = 4.08MLKK257 pKa = 10.79DD258 pKa = 3.75RR259 pKa = 11.84MVGSSKK265 pKa = 11.02YY266 pKa = 8.66EE267 pKa = 3.76SEE269 pKa = 4.18MKK271 pKa = 10.26IFRR274 pKa = 11.84HH275 pKa = 5.06NVHH278 pKa = 5.88VVVFSNEE285 pKa = 4.05YY286 pKa = 9.55PDD288 pKa = 3.95EE289 pKa = 4.37TKK291 pKa = 8.11MTADD295 pKa = 3.83RR296 pKa = 11.84YY297 pKa = 10.78DD298 pKa = 3.2INTII302 pKa = 3.52
MM1 pKa = 8.01PSPKK5 pKa = 9.1STRR8 pKa = 11.84WCFTHH13 pKa = 6.62NNWTEE18 pKa = 3.71PVFQQWSNLLGDD30 pKa = 3.7EE31 pKa = 4.59ANVKK35 pKa = 10.37FGIIGKK41 pKa = 8.85EE42 pKa = 3.9VGEE45 pKa = 4.24QGTPHH50 pKa = 6.3LQGFLILHH58 pKa = 5.86RR59 pKa = 11.84QQRR62 pKa = 11.84LSWVRR67 pKa = 11.84RR68 pKa = 11.84HH69 pKa = 6.21FPDD72 pKa = 4.16GCHH75 pKa = 5.59WSIARR80 pKa = 11.84SDD82 pKa = 3.77SEE84 pKa = 4.64TNRR87 pKa = 11.84TYY89 pKa = 10.8CKK91 pKa = 10.14KK92 pKa = 10.66DD93 pKa = 2.75GDD95 pKa = 4.2FIEE98 pKa = 5.64FGVFPDD104 pKa = 3.83AQGKK108 pKa = 7.76RR109 pKa = 11.84TDD111 pKa = 3.61LDD113 pKa = 4.1QFIEE117 pKa = 4.01WLDD120 pKa = 3.59EE121 pKa = 4.24FEE123 pKa = 4.99SNNGRR128 pKa = 11.84PASSPEE134 pKa = 3.53IAKK137 pKa = 7.36THH139 pKa = 5.86PKK141 pKa = 9.64MYY143 pKa = 10.01LRR145 pKa = 11.84YY146 pKa = 8.26PRR148 pKa = 11.84SVRR151 pKa = 11.84LAKK154 pKa = 10.09RR155 pKa = 11.84RR156 pKa = 11.84CALFPVQEE164 pKa = 4.52GDD166 pKa = 3.52LNDD169 pKa = 3.53WQRR172 pKa = 11.84EE173 pKa = 4.19LEE175 pKa = 4.37EE176 pKa = 4.85KK177 pKa = 10.8LGADD181 pKa = 3.34PDD183 pKa = 3.95DD184 pKa = 3.79RR185 pKa = 11.84TVLFYY190 pKa = 11.16VDD192 pKa = 3.62PDD194 pKa = 3.57GGKK197 pKa = 10.12GKK199 pKa = 7.61TWFVRR204 pKa = 11.84RR205 pKa = 11.84YY206 pKa = 8.26LTLHH210 pKa = 6.77PNLAQALPIGQIKK223 pKa = 10.55DD224 pKa = 3.03MAYY227 pKa = 10.29AVDD230 pKa = 3.42TNARR234 pKa = 11.84VFFVNVARR242 pKa = 11.84SGMEE246 pKa = 3.38FLPYY250 pKa = 10.26RR251 pKa = 11.84FLEE254 pKa = 4.08MLKK257 pKa = 10.79DD258 pKa = 3.75RR259 pKa = 11.84MVGSSKK265 pKa = 11.02YY266 pKa = 8.66EE267 pKa = 3.76SEE269 pKa = 4.18MKK271 pKa = 10.26IFRR274 pKa = 11.84HH275 pKa = 5.06NVHH278 pKa = 5.88VVVFSNEE285 pKa = 4.05YY286 pKa = 9.55PDD288 pKa = 3.95EE289 pKa = 4.37TKK291 pKa = 8.11MTADD295 pKa = 3.83RR296 pKa = 11.84YY297 pKa = 10.78DD298 pKa = 3.2INTII302 pKa = 3.52
Molecular weight: 35.25 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A141MJA8|A0A141MJA8_9VIRU Uncharacterized protein OS=Ctenophore-associated circular virus 2 OX=1778559 PE=4 SV=1
MM1 pKa = 7.12IQRR4 pKa = 11.84YY5 pKa = 6.04WRR7 pKa = 11.84GRR9 pKa = 11.84RR10 pKa = 11.84NRR12 pKa = 11.84HH13 pKa = 4.07QARR16 pKa = 11.84ARR18 pKa = 11.84AQIGEE23 pKa = 4.15RR24 pKa = 11.84RR25 pKa = 11.84GTATAKK31 pKa = 10.12KK32 pKa = 9.46HH33 pKa = 6.28AISDD37 pKa = 4.04TDD39 pKa = 3.31PTAYY43 pKa = 8.14GTRR46 pKa = 11.84VLNSVEE52 pKa = 3.9LTAIPSQQNQGSVDD66 pKa = 2.87IDD68 pKa = 3.55LRR70 pKa = 11.84SRR72 pKa = 11.84DD73 pKa = 3.41LAYY76 pKa = 9.82INGWKK81 pKa = 10.1FCIEE85 pKa = 3.93LRR87 pKa = 11.84NNLTEE92 pKa = 3.96PLYY95 pKa = 10.66INLAVVVPKK104 pKa = 10.61HH105 pKa = 5.84NATIDD110 pKa = 3.42PNIWFRR116 pKa = 11.84GYY118 pKa = 11.04GIDD121 pKa = 4.21RR122 pKa = 11.84AIAFDD127 pKa = 4.24PSTLNSNDD135 pKa = 3.18FHH137 pKa = 7.01CRR139 pKa = 11.84PLNPDD144 pKa = 3.44AYY146 pKa = 10.26HH147 pKa = 7.44ILMHH151 pKa = 6.38KK152 pKa = 10.12RR153 pKa = 11.84YY154 pKa = 9.37RR155 pKa = 11.84LNGDD159 pKa = 3.27TDD161 pKa = 3.84TATYY165 pKa = 10.27AHH167 pKa = 6.45NSGRR171 pKa = 11.84NFMNLNLWVPLRR183 pKa = 11.84RR184 pKa = 11.84QIRR187 pKa = 11.84FDD189 pKa = 3.34EE190 pKa = 4.45STGTGAQNGRR200 pKa = 11.84CFFVWWCDD208 pKa = 2.94KK209 pKa = 11.11YY210 pKa = 10.83MSNTGTVTTAASLQMSKK227 pKa = 10.82RR228 pKa = 11.84IICYY232 pKa = 9.86FRR234 pKa = 11.84DD235 pKa = 3.51PRR237 pKa = 11.84NN238 pKa = 3.11
MM1 pKa = 7.12IQRR4 pKa = 11.84YY5 pKa = 6.04WRR7 pKa = 11.84GRR9 pKa = 11.84RR10 pKa = 11.84NRR12 pKa = 11.84HH13 pKa = 4.07QARR16 pKa = 11.84ARR18 pKa = 11.84AQIGEE23 pKa = 4.15RR24 pKa = 11.84RR25 pKa = 11.84GTATAKK31 pKa = 10.12KK32 pKa = 9.46HH33 pKa = 6.28AISDD37 pKa = 4.04TDD39 pKa = 3.31PTAYY43 pKa = 8.14GTRR46 pKa = 11.84VLNSVEE52 pKa = 3.9LTAIPSQQNQGSVDD66 pKa = 2.87IDD68 pKa = 3.55LRR70 pKa = 11.84SRR72 pKa = 11.84DD73 pKa = 3.41LAYY76 pKa = 9.82INGWKK81 pKa = 10.1FCIEE85 pKa = 3.93LRR87 pKa = 11.84NNLTEE92 pKa = 3.96PLYY95 pKa = 10.66INLAVVVPKK104 pKa = 10.61HH105 pKa = 5.84NATIDD110 pKa = 3.42PNIWFRR116 pKa = 11.84GYY118 pKa = 11.04GIDD121 pKa = 4.21RR122 pKa = 11.84AIAFDD127 pKa = 4.24PSTLNSNDD135 pKa = 3.18FHH137 pKa = 7.01CRR139 pKa = 11.84PLNPDD144 pKa = 3.44AYY146 pKa = 10.26HH147 pKa = 7.44ILMHH151 pKa = 6.38KK152 pKa = 10.12RR153 pKa = 11.84YY154 pKa = 9.37RR155 pKa = 11.84LNGDD159 pKa = 3.27TDD161 pKa = 3.84TATYY165 pKa = 10.27AHH167 pKa = 6.45NSGRR171 pKa = 11.84NFMNLNLWVPLRR183 pKa = 11.84RR184 pKa = 11.84QIRR187 pKa = 11.84FDD189 pKa = 3.34EE190 pKa = 4.45STGTGAQNGRR200 pKa = 11.84CFFVWWCDD208 pKa = 2.94KK209 pKa = 11.11YY210 pKa = 10.83MSNTGTVTTAASLQMSKK227 pKa = 10.82RR228 pKa = 11.84IICYY232 pKa = 9.86FRR234 pKa = 11.84DD235 pKa = 3.51PRR237 pKa = 11.84NN238 pKa = 3.11
Molecular weight: 27.49 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
540 |
238 |
302 |
270.0 |
31.37 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.296 ± 1.083 | 1.667 ± 0.279 |
6.852 ± 0.353 | 4.63 ± 1.624 |
5.185 ± 0.902 | 6.111 ± 0.147 |
2.963 ± 0.014 | 5.185 ± 0.988 |
4.444 ± 0.966 | 7.037 ± 0.202 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.407 ± 0.197 | 6.296 ± 1.353 |
5.0 ± 0.513 | 3.889 ± 0.069 |
9.259 ± 1.069 | 5.185 ± 0.092 |
6.111 ± 0.933 | 5.185 ± 0.902 |
2.593 ± 0.046 | 3.704 ± 0.32 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
