
Neolentinus lepideus HHB14362 ss-1
Taxonomy: cellular organisms; Eukaryota; Opisthokonta; Fungi; Dikarya; Basidiomycota; Agaricomycotina; Agaricomycetes; Agaricomycetes incertae sedis; Gloeophyllales; Gloeophyllaceae; Neolentinus; Neolentinus lepideus
Average proteome isoelectric point is 6.69
Get precalculated fractions of proteins
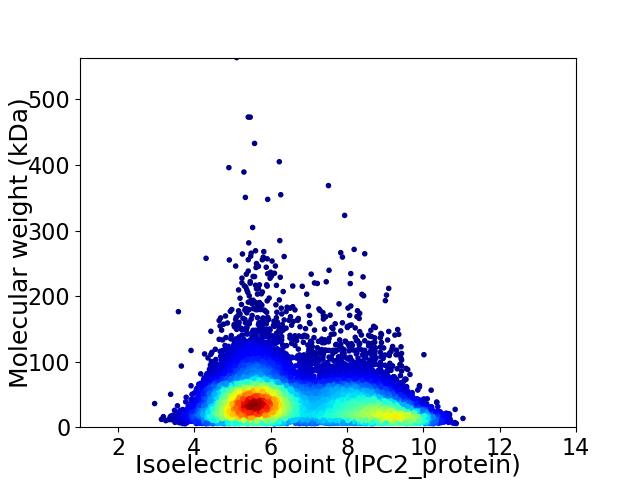
Virtual 2D-PAGE plot for 13108 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A165Q383|A0A165Q383_9AGAM Uncharacterized protein OS=Neolentinus lepideus HHB14362 ss-1 OX=1314782 GN=NEOLEDRAFT_1171731 PE=4 SV=1
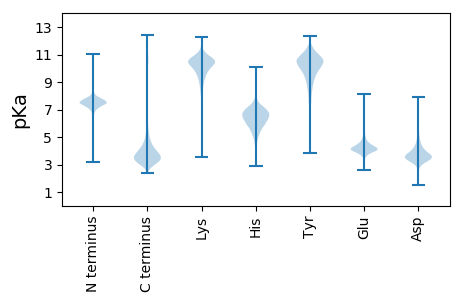
MM1 pKa = 7.37SFCVDD6 pKa = 3.89PSTTFVDD13 pKa = 3.5QTLVSDD19 pKa = 3.83IQAPIFSTHH28 pKa = 5.83VDD30 pKa = 3.56CADD33 pKa = 3.48AGAAPSVHH41 pKa = 6.64IDD43 pKa = 3.58LAAFNTSAKK52 pKa = 8.94VNSMSHH58 pKa = 5.93LTTNGASEE66 pKa = 4.38NVSFPAPSLRR76 pKa = 11.84DD77 pKa = 3.5EE78 pKa = 4.51VFLPSTFTGGYY89 pKa = 6.88WRR91 pKa = 11.84SHH93 pKa = 4.46EE94 pKa = 4.28VGVCVDD100 pKa = 3.19GTEE103 pKa = 4.7RR104 pKa = 11.84SLEE107 pKa = 4.05DD108 pKa = 3.39LAAVASLGDD117 pKa = 3.7QPDD120 pKa = 3.52VAVFGDD126 pKa = 3.62ARR128 pKa = 11.84RR129 pKa = 11.84NGPMSCSAPVDD140 pKa = 3.34QLQQLEE146 pKa = 4.24QDD148 pKa = 4.14FRR150 pKa = 11.84AQPAINGCAPLGPDD164 pKa = 4.17AADD167 pKa = 3.42ASAPPSTPLLAMKK180 pKa = 10.11ALPAEE185 pKa = 4.6DD186 pKa = 4.47VVDD189 pKa = 3.96DD190 pKa = 4.68HH191 pKa = 7.26PSFVDD196 pKa = 4.13DD197 pKa = 4.3SQSSTWLSFEE207 pKa = 4.26HH208 pKa = 7.22ADD210 pKa = 4.27VDD212 pKa = 4.79LEE214 pKa = 4.6LPALLDD220 pKa = 3.78SCDD223 pKa = 3.53VPFNSPSPSYY233 pKa = 11.01NIGLFDD239 pKa = 4.64FDD241 pKa = 4.87YY242 pKa = 10.82PPSPSTASATSSFDD256 pKa = 3.61SPPPTPLLSSLDD268 pKa = 3.79ISCLEE273 pKa = 4.16CSQYY277 pKa = 11.37AQDD280 pKa = 3.43GHH282 pKa = 6.55AVCGTCGFTALLGMEE297 pKa = 4.94GGCGGSGVRR306 pKa = 11.84VVNAGEE312 pKa = 4.02DD313 pKa = 3.59LMDD316 pKa = 3.84GVEE319 pKa = 4.37DD320 pKa = 4.77SRR322 pKa = 11.84GGCCDD327 pKa = 4.07VIPGLQHH334 pKa = 5.91TWTSKK339 pKa = 9.87IPLDD343 pKa = 3.69GGYY346 pKa = 10.62KK347 pKa = 10.4FGQLPPQAFEE357 pKa = 3.78QDD359 pKa = 3.47EE360 pKa = 4.78VNTLGVDD367 pKa = 4.67FSAA370 pKa = 5.42
MM1 pKa = 7.37SFCVDD6 pKa = 3.89PSTTFVDD13 pKa = 3.5QTLVSDD19 pKa = 3.83IQAPIFSTHH28 pKa = 5.83VDD30 pKa = 3.56CADD33 pKa = 3.48AGAAPSVHH41 pKa = 6.64IDD43 pKa = 3.58LAAFNTSAKK52 pKa = 8.94VNSMSHH58 pKa = 5.93LTTNGASEE66 pKa = 4.38NVSFPAPSLRR76 pKa = 11.84DD77 pKa = 3.5EE78 pKa = 4.51VFLPSTFTGGYY89 pKa = 6.88WRR91 pKa = 11.84SHH93 pKa = 4.46EE94 pKa = 4.28VGVCVDD100 pKa = 3.19GTEE103 pKa = 4.7RR104 pKa = 11.84SLEE107 pKa = 4.05DD108 pKa = 3.39LAAVASLGDD117 pKa = 3.7QPDD120 pKa = 3.52VAVFGDD126 pKa = 3.62ARR128 pKa = 11.84RR129 pKa = 11.84NGPMSCSAPVDD140 pKa = 3.34QLQQLEE146 pKa = 4.24QDD148 pKa = 4.14FRR150 pKa = 11.84AQPAINGCAPLGPDD164 pKa = 4.17AADD167 pKa = 3.42ASAPPSTPLLAMKK180 pKa = 10.11ALPAEE185 pKa = 4.6DD186 pKa = 4.47VVDD189 pKa = 3.96DD190 pKa = 4.68HH191 pKa = 7.26PSFVDD196 pKa = 4.13DD197 pKa = 4.3SQSSTWLSFEE207 pKa = 4.26HH208 pKa = 7.22ADD210 pKa = 4.27VDD212 pKa = 4.79LEE214 pKa = 4.6LPALLDD220 pKa = 3.78SCDD223 pKa = 3.53VPFNSPSPSYY233 pKa = 11.01NIGLFDD239 pKa = 4.64FDD241 pKa = 4.87YY242 pKa = 10.82PPSPSTASATSSFDD256 pKa = 3.61SPPPTPLLSSLDD268 pKa = 3.79ISCLEE273 pKa = 4.16CSQYY277 pKa = 11.37AQDD280 pKa = 3.43GHH282 pKa = 6.55AVCGTCGFTALLGMEE297 pKa = 4.94GGCGGSGVRR306 pKa = 11.84VVNAGEE312 pKa = 4.02DD313 pKa = 3.59LMDD316 pKa = 3.84GVEE319 pKa = 4.37DD320 pKa = 4.77SRR322 pKa = 11.84GGCCDD327 pKa = 4.07VIPGLQHH334 pKa = 5.91TWTSKK339 pKa = 9.87IPLDD343 pKa = 3.69GGYY346 pKa = 10.62KK347 pKa = 10.4FGQLPPQAFEE357 pKa = 3.78QDD359 pKa = 3.47EE360 pKa = 4.78VNTLGVDD367 pKa = 4.67FSAA370 pKa = 5.42
Molecular weight: 38.68 kDa
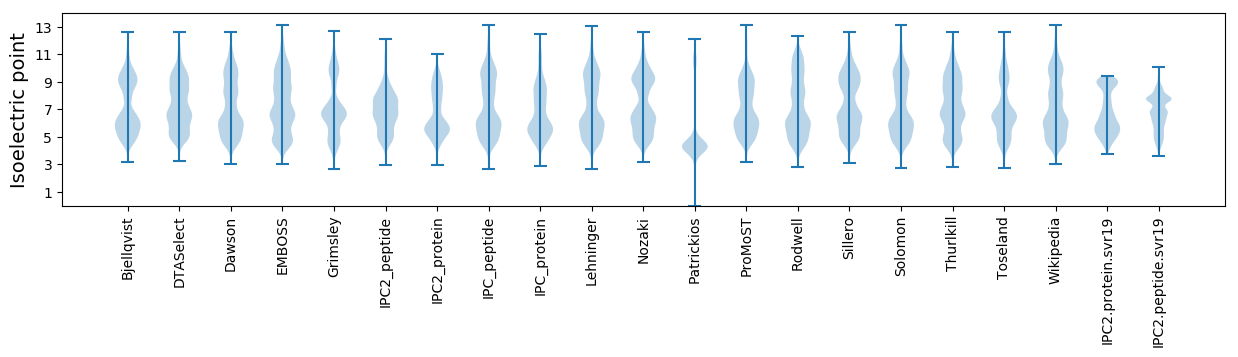
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A165NQK6|A0A165NQK6_9AGAM G-patch domain-containing protein OS=Neolentinus lepideus HHB14362 ss-1 OX=1314782 GN=NEOLEDRAFT_1182897 PE=4 SV=1
MM1 pKa = 7.54PRR3 pKa = 11.84ILKK6 pKa = 9.67QVAQLLARR14 pKa = 11.84PPALPHH20 pKa = 6.24ASSVVSTLTGLHH32 pKa = 6.2RR33 pKa = 11.84PSSQRR38 pKa = 11.84LTPLPFALAFQSSSLLTPSFVGRR61 pKa = 11.84SSALLQLNQVRR72 pKa = 11.84WGSRR76 pKa = 11.84GTEE79 pKa = 3.93YY80 pKa = 10.77QPSQRR85 pKa = 11.84VRR87 pKa = 11.84KK88 pKa = 9.28RR89 pKa = 11.84RR90 pKa = 11.84HH91 pKa = 4.87GFLARR96 pKa = 11.84KK97 pKa = 9.53KK98 pKa = 9.03SQHH101 pKa = 5.36GPKK104 pKa = 9.63ILARR108 pKa = 11.84RR109 pKa = 11.84RR110 pKa = 11.84AKK112 pKa = 9.85GRR114 pKa = 11.84KK115 pKa = 8.43FLSHH119 pKa = 6.98
MM1 pKa = 7.54PRR3 pKa = 11.84ILKK6 pKa = 9.67QVAQLLARR14 pKa = 11.84PPALPHH20 pKa = 6.24ASSVVSTLTGLHH32 pKa = 6.2RR33 pKa = 11.84PSSQRR38 pKa = 11.84LTPLPFALAFQSSSLLTPSFVGRR61 pKa = 11.84SSALLQLNQVRR72 pKa = 11.84WGSRR76 pKa = 11.84GTEE79 pKa = 3.93YY80 pKa = 10.77QPSQRR85 pKa = 11.84VRR87 pKa = 11.84KK88 pKa = 9.28RR89 pKa = 11.84RR90 pKa = 11.84HH91 pKa = 4.87GFLARR96 pKa = 11.84KK97 pKa = 9.53KK98 pKa = 9.03SQHH101 pKa = 5.36GPKK104 pKa = 9.63ILARR108 pKa = 11.84RR109 pKa = 11.84RR110 pKa = 11.84AKK112 pKa = 9.85GRR114 pKa = 11.84KK115 pKa = 8.43FLSHH119 pKa = 6.98
Molecular weight: 13.35 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
5153932 |
49 |
5057 |
393.2 |
43.53 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
8.431 ± 0.019 | 1.331 ± 0.009 |
5.557 ± 0.013 | 6.016 ± 0.021 |
3.597 ± 0.013 | 6.504 ± 0.021 |
2.525 ± 0.01 | 4.819 ± 0.016 |
4.535 ± 0.02 | 9.236 ± 0.027 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.181 ± 0.008 | 3.301 ± 0.01 |
6.398 ± 0.025 | 3.681 ± 0.014 |
6.362 ± 0.021 | 8.743 ± 0.03 |
5.935 ± 0.015 | 6.554 ± 0.016 |
1.496 ± 0.009 | 2.799 ± 0.012 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
