
Wuhan pillworm virus 3
Taxonomy: Viruses; Riboviria; unclassified Riboviria; unclassified RNA viruses ShiM-2016
Average proteome isoelectric point is 7.98
Get precalculated fractions of proteins

Virtual 2D-PAGE plot for 3 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions

Protein with the lowest isoelectric point:
>tr|A0A1L3KF15|A0A1L3KF15_9VIRU RNA-directed RNA polymerase OS=Wuhan pillworm virus 3 OX=1923746 PE=4 SV=1
MM1 pKa = 8.0RR2 pKa = 11.84DD3 pKa = 3.36SDD5 pKa = 4.5PPSKK9 pKa = 10.84DD10 pKa = 2.74IARR13 pKa = 11.84EE14 pKa = 3.82ALFRR18 pKa = 11.84TLSDD22 pKa = 3.4FRR24 pKa = 11.84HH25 pKa = 5.75LQRR28 pKa = 11.84DD29 pKa = 4.31CDD31 pKa = 4.07PEE33 pKa = 4.25WYY35 pKa = 10.08SDD37 pKa = 3.36EE38 pKa = 4.72NMDD41 pKa = 5.26RR42 pKa = 11.84VINSLNRR49 pKa = 11.84NASPGNPFCWVANNGQLIDD68 pKa = 3.53KK69 pKa = 10.28HH70 pKa = 7.49RR71 pKa = 11.84EE72 pKa = 3.74LLKK75 pKa = 9.68TLVRR79 pKa = 11.84QRR81 pKa = 11.84LNGASAYY88 pKa = 9.36PIRR91 pKa = 11.84LFVKK95 pKa = 9.98PEE97 pKa = 3.41WHH99 pKa = 6.56KK100 pKa = 10.79KK101 pKa = 9.48SKK103 pKa = 10.39IEE105 pKa = 3.77EE106 pKa = 3.85GRR108 pKa = 11.84YY109 pKa = 8.89RR110 pKa = 11.84LIWSVSVVDD119 pKa = 4.76QIIDD123 pKa = 3.35KK124 pKa = 9.87MLLGDD129 pKa = 4.53YY130 pKa = 10.68LDD132 pKa = 4.22QEE134 pKa = 4.17PDD136 pKa = 2.48NWIFHH141 pKa = 6.12PSKK144 pKa = 11.02VGWSYY149 pKa = 11.81LKK151 pKa = 10.92GGWKK155 pKa = 9.76YY156 pKa = 10.1VPKK159 pKa = 10.78GVGTDD164 pKa = 4.13FSAWDD169 pKa = 3.4MTAQQWLLDD178 pKa = 4.19LLCEE182 pKa = 4.7FYY184 pKa = 10.68INQHH188 pKa = 6.09RR189 pKa = 11.84NPTAYY194 pKa = 8.86WISWVRR200 pKa = 11.84RR201 pKa = 11.84RR202 pKa = 11.84FGEE205 pKa = 4.35LYY207 pKa = 10.55HH208 pKa = 7.2GGALVQLSDD217 pKa = 3.64GTLLEE222 pKa = 4.01QLFGGIQKK230 pKa = 10.56SGTLYY235 pKa = 9.89TLSGNSIMQVILHH248 pKa = 5.95HH249 pKa = 6.19VICLSLNYY257 pKa = 10.46DD258 pKa = 3.68SVPWLWVLGDD268 pKa = 4.28DD269 pKa = 4.4RR270 pKa = 11.84LQEE273 pKa = 4.26RR274 pKa = 11.84VDD276 pKa = 3.63KK277 pKa = 10.75EE278 pKa = 3.86YY279 pKa = 11.52ADD281 pKa = 3.62ALKK284 pKa = 10.44RR285 pKa = 11.84WVVLKK290 pKa = 10.54DD291 pKa = 2.74ISYY294 pKa = 11.02NEE296 pKa = 4.35FCGMHH301 pKa = 6.03YY302 pKa = 10.81GSTIEE307 pKa = 3.48PAYY310 pKa = 10.27RR311 pKa = 11.84SKK313 pKa = 11.16HH314 pKa = 5.39LLNLAADD321 pKa = 4.15PTPEE325 pKa = 4.05RR326 pKa = 11.84LMSYY330 pKa = 9.99QLLYY334 pKa = 10.27RR335 pKa = 11.84HH336 pKa = 6.15SKK338 pKa = 10.2FYY340 pKa = 9.9PALVRR345 pKa = 11.84FIRR348 pKa = 11.84SLNVQPAHH356 pKa = 6.5EE357 pKa = 4.19IWVDD361 pKa = 3.98EE362 pKa = 4.29VWGG365 pKa = 3.85
MM1 pKa = 8.0RR2 pKa = 11.84DD3 pKa = 3.36SDD5 pKa = 4.5PPSKK9 pKa = 10.84DD10 pKa = 2.74IARR13 pKa = 11.84EE14 pKa = 3.82ALFRR18 pKa = 11.84TLSDD22 pKa = 3.4FRR24 pKa = 11.84HH25 pKa = 5.75LQRR28 pKa = 11.84DD29 pKa = 4.31CDD31 pKa = 4.07PEE33 pKa = 4.25WYY35 pKa = 10.08SDD37 pKa = 3.36EE38 pKa = 4.72NMDD41 pKa = 5.26RR42 pKa = 11.84VINSLNRR49 pKa = 11.84NASPGNPFCWVANNGQLIDD68 pKa = 3.53KK69 pKa = 10.28HH70 pKa = 7.49RR71 pKa = 11.84EE72 pKa = 3.74LLKK75 pKa = 9.68TLVRR79 pKa = 11.84QRR81 pKa = 11.84LNGASAYY88 pKa = 9.36PIRR91 pKa = 11.84LFVKK95 pKa = 9.98PEE97 pKa = 3.41WHH99 pKa = 6.56KK100 pKa = 10.79KK101 pKa = 9.48SKK103 pKa = 10.39IEE105 pKa = 3.77EE106 pKa = 3.85GRR108 pKa = 11.84YY109 pKa = 8.89RR110 pKa = 11.84LIWSVSVVDD119 pKa = 4.76QIIDD123 pKa = 3.35KK124 pKa = 9.87MLLGDD129 pKa = 4.53YY130 pKa = 10.68LDD132 pKa = 4.22QEE134 pKa = 4.17PDD136 pKa = 2.48NWIFHH141 pKa = 6.12PSKK144 pKa = 11.02VGWSYY149 pKa = 11.81LKK151 pKa = 10.92GGWKK155 pKa = 9.76YY156 pKa = 10.1VPKK159 pKa = 10.78GVGTDD164 pKa = 4.13FSAWDD169 pKa = 3.4MTAQQWLLDD178 pKa = 4.19LLCEE182 pKa = 4.7FYY184 pKa = 10.68INQHH188 pKa = 6.09RR189 pKa = 11.84NPTAYY194 pKa = 8.86WISWVRR200 pKa = 11.84RR201 pKa = 11.84RR202 pKa = 11.84FGEE205 pKa = 4.35LYY207 pKa = 10.55HH208 pKa = 7.2GGALVQLSDD217 pKa = 3.64GTLLEE222 pKa = 4.01QLFGGIQKK230 pKa = 10.56SGTLYY235 pKa = 9.89TLSGNSIMQVILHH248 pKa = 5.95HH249 pKa = 6.19VICLSLNYY257 pKa = 10.46DD258 pKa = 3.68SVPWLWVLGDD268 pKa = 4.28DD269 pKa = 4.4RR270 pKa = 11.84LQEE273 pKa = 4.26RR274 pKa = 11.84VDD276 pKa = 3.63KK277 pKa = 10.75EE278 pKa = 3.86YY279 pKa = 11.52ADD281 pKa = 3.62ALKK284 pKa = 10.44RR285 pKa = 11.84WVVLKK290 pKa = 10.54DD291 pKa = 2.74ISYY294 pKa = 11.02NEE296 pKa = 4.35FCGMHH301 pKa = 6.03YY302 pKa = 10.81GSTIEE307 pKa = 3.48PAYY310 pKa = 10.27RR311 pKa = 11.84SKK313 pKa = 11.16HH314 pKa = 5.39LLNLAADD321 pKa = 4.15PTPEE325 pKa = 4.05RR326 pKa = 11.84LMSYY330 pKa = 9.99QLLYY334 pKa = 10.27RR335 pKa = 11.84HH336 pKa = 6.15SKK338 pKa = 10.2FYY340 pKa = 9.9PALVRR345 pKa = 11.84FIRR348 pKa = 11.84SLNVQPAHH356 pKa = 6.5EE357 pKa = 4.19IWVDD361 pKa = 3.98EE362 pKa = 4.29VWGG365 pKa = 3.85
Molecular weight: 42.74 kDa
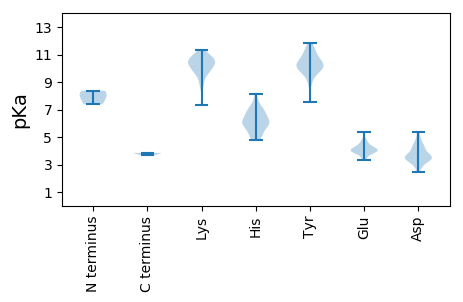
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1L3KF15|A0A1L3KF15_9VIRU RNA-directed RNA polymerase OS=Wuhan pillworm virus 3 OX=1923746 PE=4 SV=1
MM1 pKa = 7.38NAQTKK6 pKa = 10.29KK7 pKa = 8.92ITCHH11 pKa = 5.17ACGRR15 pKa = 11.84RR16 pKa = 11.84FNTQQALHH24 pKa = 5.15QHH26 pKa = 4.8QQAVHH31 pKa = 6.07GAANQRR37 pKa = 11.84VPRR40 pKa = 11.84GAGRR44 pKa = 11.84GGRR47 pKa = 11.84GRR49 pKa = 11.84RR50 pKa = 11.84NGQNNQGPARR60 pKa = 11.84QGTAQSVTSRR70 pKa = 11.84ATPNQGVSVSGFEE83 pKa = 4.07RR84 pKa = 11.84LGSIDD89 pKa = 3.22IVNAASASWQVNAWMTPRR107 pKa = 11.84LQAMSSAFQRR117 pKa = 11.84IHH119 pKa = 5.95YY120 pKa = 10.12LSLKK124 pKa = 9.14VNVVAFGSAIGTGGYY139 pKa = 10.37VIGFVADD146 pKa = 4.17PSDD149 pKa = 3.75ATPTISQLQSQAGARR164 pKa = 11.84TCKK167 pKa = 10.05SWEE170 pKa = 4.04NCDD173 pKa = 3.38VPLVVPGTLYY183 pKa = 9.48YY184 pKa = 10.2TSRR187 pKa = 11.84DD188 pKa = 3.61NEE190 pKa = 4.11PRR192 pKa = 11.84WYY194 pKa = 10.01SPGRR198 pKa = 11.84IVILVDD204 pKa = 4.9GIASAGGKK212 pKa = 9.59LVINLHH218 pKa = 5.08WSVRR222 pKa = 11.84LSVPTATPPTVLKK235 pKa = 10.78DD236 pKa = 3.29IFLSEE241 pKa = 4.28HH242 pKa = 6.79LYY244 pKa = 10.73VIPGKK249 pKa = 10.39NILGWKK255 pKa = 10.12DD256 pKa = 3.03GTTFRR261 pKa = 11.84GNISKK266 pKa = 10.16FLSTPVPGDD275 pKa = 2.79SWLSVPTFGIEE286 pKa = 3.98YY287 pKa = 10.22NEE289 pKa = 4.4GVGDD293 pKa = 3.57TGTILIHH300 pKa = 4.89YY301 pKa = 7.62LKK303 pKa = 10.32YY304 pKa = 10.52NSSEE308 pKa = 4.01GSLTCSNDD316 pKa = 3.06GEE318 pKa = 4.49KK319 pKa = 10.75VVTQVWQASVAAQVVVPRR337 pKa = 11.84NTILTIVDD345 pKa = 3.56YY346 pKa = 11.19NKK348 pKa = 9.44PAKK351 pKa = 9.75PAVSDD356 pKa = 4.41FIQPLVSNFNNVGIDD371 pKa = 3.31SSRR374 pKa = 11.84NSTTPSAGEE383 pKa = 4.1TGTDD387 pKa = 3.63PPYY390 pKa = 10.2STYY393 pKa = 11.26LEE395 pKa = 4.05LLRR398 pKa = 11.84RR399 pKa = 11.84QSALDD404 pKa = 3.54SKK406 pKa = 10.73ISRR409 pKa = 11.84IEE411 pKa = 3.87LQLSKK416 pKa = 9.99MVKK419 pKa = 9.83VLMPKK424 pKa = 10.38DD425 pKa = 3.56QDD427 pKa = 3.54SSQSSRR433 pKa = 11.84SRR435 pKa = 11.84SPSKK439 pKa = 10.45DD440 pKa = 3.01TTKK443 pKa = 10.77LDD445 pKa = 3.77SKK447 pKa = 10.8GTT449 pKa = 3.68
MM1 pKa = 7.38NAQTKK6 pKa = 10.29KK7 pKa = 8.92ITCHH11 pKa = 5.17ACGRR15 pKa = 11.84RR16 pKa = 11.84FNTQQALHH24 pKa = 5.15QHH26 pKa = 4.8QQAVHH31 pKa = 6.07GAANQRR37 pKa = 11.84VPRR40 pKa = 11.84GAGRR44 pKa = 11.84GGRR47 pKa = 11.84GRR49 pKa = 11.84RR50 pKa = 11.84NGQNNQGPARR60 pKa = 11.84QGTAQSVTSRR70 pKa = 11.84ATPNQGVSVSGFEE83 pKa = 4.07RR84 pKa = 11.84LGSIDD89 pKa = 3.22IVNAASASWQVNAWMTPRR107 pKa = 11.84LQAMSSAFQRR117 pKa = 11.84IHH119 pKa = 5.95YY120 pKa = 10.12LSLKK124 pKa = 9.14VNVVAFGSAIGTGGYY139 pKa = 10.37VIGFVADD146 pKa = 4.17PSDD149 pKa = 3.75ATPTISQLQSQAGARR164 pKa = 11.84TCKK167 pKa = 10.05SWEE170 pKa = 4.04NCDD173 pKa = 3.38VPLVVPGTLYY183 pKa = 9.48YY184 pKa = 10.2TSRR187 pKa = 11.84DD188 pKa = 3.61NEE190 pKa = 4.11PRR192 pKa = 11.84WYY194 pKa = 10.01SPGRR198 pKa = 11.84IVILVDD204 pKa = 4.9GIASAGGKK212 pKa = 9.59LVINLHH218 pKa = 5.08WSVRR222 pKa = 11.84LSVPTATPPTVLKK235 pKa = 10.78DD236 pKa = 3.29IFLSEE241 pKa = 4.28HH242 pKa = 6.79LYY244 pKa = 10.73VIPGKK249 pKa = 10.39NILGWKK255 pKa = 10.12DD256 pKa = 3.03GTTFRR261 pKa = 11.84GNISKK266 pKa = 10.16FLSTPVPGDD275 pKa = 2.79SWLSVPTFGIEE286 pKa = 3.98YY287 pKa = 10.22NEE289 pKa = 4.4GVGDD293 pKa = 3.57TGTILIHH300 pKa = 4.89YY301 pKa = 7.62LKK303 pKa = 10.32YY304 pKa = 10.52NSSEE308 pKa = 4.01GSLTCSNDD316 pKa = 3.06GEE318 pKa = 4.49KK319 pKa = 10.75VVTQVWQASVAAQVVVPRR337 pKa = 11.84NTILTIVDD345 pKa = 3.56YY346 pKa = 11.19NKK348 pKa = 9.44PAKK351 pKa = 9.75PAVSDD356 pKa = 4.41FIQPLVSNFNNVGIDD371 pKa = 3.31SSRR374 pKa = 11.84NSTTPSAGEE383 pKa = 4.1TGTDD387 pKa = 3.63PPYY390 pKa = 10.2STYY393 pKa = 11.26LEE395 pKa = 4.05LLRR398 pKa = 11.84RR399 pKa = 11.84QSALDD404 pKa = 3.54SKK406 pKa = 10.73ISRR409 pKa = 11.84IEE411 pKa = 3.87LQLSKK416 pKa = 9.99MVKK419 pKa = 9.83VLMPKK424 pKa = 10.38DD425 pKa = 3.56QDD427 pKa = 3.54SSQSSRR433 pKa = 11.84SRR435 pKa = 11.84SPSKK439 pKa = 10.45DD440 pKa = 3.01TTKK443 pKa = 10.77LDD445 pKa = 3.77SKK447 pKa = 10.8GTT449 pKa = 3.68
Molecular weight: 48.49 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1319 |
365 |
505 |
439.7 |
49.51 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
5.534 ± 0.697 | 1.061 ± 0.14 |
5.914 ± 0.646 | 3.563 ± 0.552 |
3.487 ± 0.573 | 7.354 ± 0.611 |
2.35 ± 0.339 | 5.838 ± 0.44 |
5.459 ± 0.647 | 9.249 ± 1.103 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.592 ± 0.209 | 4.397 ± 0.479 |
5.004 ± 0.349 | 5.08 ± 0.373 |
5.838 ± 0.398 | 9.325 ± 0.841 |
5.762 ± 1.09 | 6.596 ± 0.894 |
2.957 ± 0.58 | 3.639 ± 0.501 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
