
Phenylobacterium parvum
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Alphaproteobacteria; Caulobacterales; Caulobacteraceae; Phenylobacterium
Average proteome isoelectric point is 6.55
Get precalculated fractions of proteins
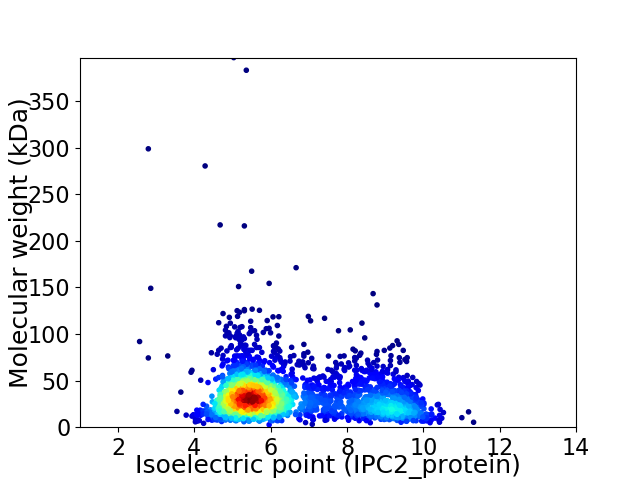
Virtual 2D-PAGE plot for 2596 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A2Z3HNY5|A0A2Z3HNY5_9CAUL DUF1289 domain-containing protein OS=Phenylobacterium parvum OX=2201350 GN=HYN04_04015 PE=4 SV=1
MM1 pKa = 6.59GTYY4 pKa = 10.08YY5 pKa = 10.88YY6 pKa = 10.72AGHH9 pKa = 7.17TYY11 pKa = 10.41QIVKK15 pKa = 8.82TAMTWAAAKK24 pKa = 10.02AWAEE28 pKa = 4.14SQGGHH33 pKa = 6.4LAYY36 pKa = 8.99ITSAAEE42 pKa = 3.8NQALVSMTQLTTGLDD57 pKa = 4.0LAPSAADD64 pKa = 3.4GGGARR69 pKa = 11.84YY70 pKa = 8.77LWLGGSDD77 pKa = 3.72ADD79 pKa = 5.5VEE81 pKa = 4.88GTWRR85 pKa = 11.84WGDD88 pKa = 3.47GTLVTSGYY96 pKa = 9.68SNWGAGALGVEE107 pKa = 4.24PDD109 pKa = 3.7NYY111 pKa = 11.15GGVQDD116 pKa = 5.06AMAFGLQSWPQPSGGIGTAFKK137 pKa = 10.63WNDD140 pKa = 3.05INPANSLYY148 pKa = 10.57FVVEE152 pKa = 3.73WDD154 pKa = 4.21HH155 pKa = 7.8IRR157 pKa = 11.84GTTGADD163 pKa = 3.5SISQTGNATVYY174 pKa = 10.58GLEE177 pKa = 4.25GNDD180 pKa = 4.38IISLAQGSNYY190 pKa = 9.74LRR192 pKa = 11.84GDD194 pKa = 3.59AGDD197 pKa = 4.13DD198 pKa = 3.79SVFGGSGFDD207 pKa = 4.96DD208 pKa = 3.42INGNMGADD216 pKa = 3.54TLRR219 pKa = 11.84GNDD222 pKa = 3.63GEE224 pKa = 4.26DD225 pKa = 2.89WVVGGKK231 pKa = 10.32DD232 pKa = 3.05NDD234 pKa = 4.99LIFGDD239 pKa = 3.73AAFDD243 pKa = 3.41IVYY246 pKa = 10.68GNMGNDD252 pKa = 3.62TVDD255 pKa = 3.22GGAGNDD261 pKa = 3.47WVRR264 pKa = 11.84GGQGDD269 pKa = 4.02DD270 pKa = 3.76SVVSGAGDD278 pKa = 3.21DD279 pKa = 3.97WLWGDD284 pKa = 4.54RR285 pKa = 11.84GNDD288 pKa = 3.95TISGGAGADD297 pKa = 3.45LFYY300 pKa = 11.15SFAGAGIDD308 pKa = 3.86RR309 pKa = 11.84ITDD312 pKa = 3.67FSYY315 pKa = 11.63AEE317 pKa = 4.05GDD319 pKa = 3.57RR320 pKa = 11.84LKK322 pKa = 11.36LEE324 pKa = 4.64GGPSYY329 pKa = 11.06SVSQVGADD337 pKa = 3.54VIVDD341 pKa = 3.89MGNGDD346 pKa = 3.66QVILVGVSLASLGSGWILL364 pKa = 3.58
MM1 pKa = 6.59GTYY4 pKa = 10.08YY5 pKa = 10.88YY6 pKa = 10.72AGHH9 pKa = 7.17TYY11 pKa = 10.41QIVKK15 pKa = 8.82TAMTWAAAKK24 pKa = 10.02AWAEE28 pKa = 4.14SQGGHH33 pKa = 6.4LAYY36 pKa = 8.99ITSAAEE42 pKa = 3.8NQALVSMTQLTTGLDD57 pKa = 4.0LAPSAADD64 pKa = 3.4GGGARR69 pKa = 11.84YY70 pKa = 8.77LWLGGSDD77 pKa = 3.72ADD79 pKa = 5.5VEE81 pKa = 4.88GTWRR85 pKa = 11.84WGDD88 pKa = 3.47GTLVTSGYY96 pKa = 9.68SNWGAGALGVEE107 pKa = 4.24PDD109 pKa = 3.7NYY111 pKa = 11.15GGVQDD116 pKa = 5.06AMAFGLQSWPQPSGGIGTAFKK137 pKa = 10.63WNDD140 pKa = 3.05INPANSLYY148 pKa = 10.57FVVEE152 pKa = 3.73WDD154 pKa = 4.21HH155 pKa = 7.8IRR157 pKa = 11.84GTTGADD163 pKa = 3.5SISQTGNATVYY174 pKa = 10.58GLEE177 pKa = 4.25GNDD180 pKa = 4.38IISLAQGSNYY190 pKa = 9.74LRR192 pKa = 11.84GDD194 pKa = 3.59AGDD197 pKa = 4.13DD198 pKa = 3.79SVFGGSGFDD207 pKa = 4.96DD208 pKa = 3.42INGNMGADD216 pKa = 3.54TLRR219 pKa = 11.84GNDD222 pKa = 3.63GEE224 pKa = 4.26DD225 pKa = 2.89WVVGGKK231 pKa = 10.32DD232 pKa = 3.05NDD234 pKa = 4.99LIFGDD239 pKa = 3.73AAFDD243 pKa = 3.41IVYY246 pKa = 10.68GNMGNDD252 pKa = 3.62TVDD255 pKa = 3.22GGAGNDD261 pKa = 3.47WVRR264 pKa = 11.84GGQGDD269 pKa = 4.02DD270 pKa = 3.76SVVSGAGDD278 pKa = 3.21DD279 pKa = 3.97WLWGDD284 pKa = 4.54RR285 pKa = 11.84GNDD288 pKa = 3.95TISGGAGADD297 pKa = 3.45LFYY300 pKa = 11.15SFAGAGIDD308 pKa = 3.86RR309 pKa = 11.84ITDD312 pKa = 3.67FSYY315 pKa = 11.63AEE317 pKa = 4.05GDD319 pKa = 3.57RR320 pKa = 11.84LKK322 pKa = 11.36LEE324 pKa = 4.64GGPSYY329 pKa = 11.06SVSQVGADD337 pKa = 3.54VIVDD341 pKa = 3.89MGNGDD346 pKa = 3.66QVILVGVSLASLGSGWILL364 pKa = 3.58
Molecular weight: 37.66 kDa
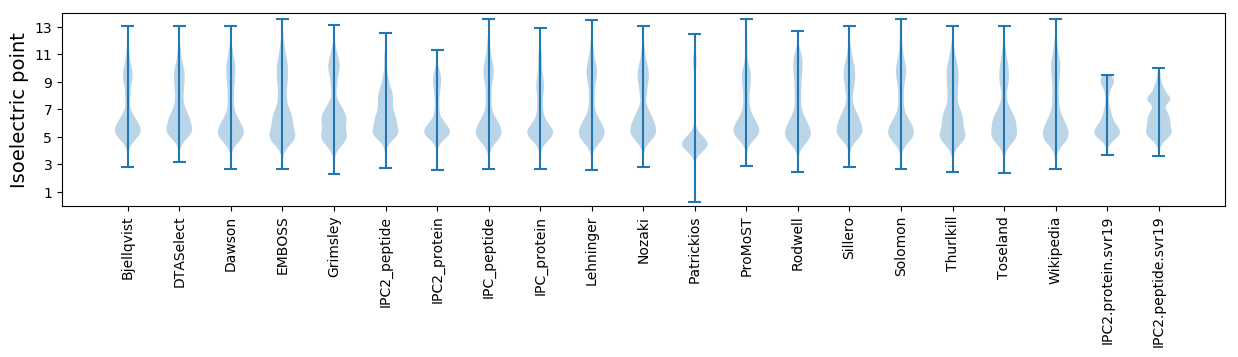
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A2Z3HYR7|A0A2Z3HYR7_9CAUL TetR/AcrR family transcriptional regulator OS=Phenylobacterium parvum OX=2201350 GN=HYN04_11110 PE=4 SV=1
MM1 pKa = 7.45KK2 pKa = 9.56RR3 pKa = 11.84TFQPSRR9 pKa = 11.84LVRR12 pKa = 11.84ARR14 pKa = 11.84RR15 pKa = 11.84HH16 pKa = 3.78GWRR19 pKa = 11.84ARR21 pKa = 11.84MATKK25 pKa = 10.5NGRR28 pKa = 11.84KK29 pKa = 9.27VIARR33 pKa = 11.84RR34 pKa = 11.84RR35 pKa = 11.84SKK37 pKa = 10.46GRR39 pKa = 11.84KK40 pKa = 8.99RR41 pKa = 11.84LTAA44 pKa = 4.18
MM1 pKa = 7.45KK2 pKa = 9.56RR3 pKa = 11.84TFQPSRR9 pKa = 11.84LVRR12 pKa = 11.84ARR14 pKa = 11.84RR15 pKa = 11.84HH16 pKa = 3.78GWRR19 pKa = 11.84ARR21 pKa = 11.84MATKK25 pKa = 10.5NGRR28 pKa = 11.84KK29 pKa = 9.27VIARR33 pKa = 11.84RR34 pKa = 11.84RR35 pKa = 11.84SKK37 pKa = 10.46GRR39 pKa = 11.84KK40 pKa = 8.99RR41 pKa = 11.84LTAA44 pKa = 4.18
Molecular weight: 5.3 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
863946 |
28 |
4068 |
332.8 |
35.66 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
13.986 ± 0.067 | 0.765 ± 0.015 |
5.745 ± 0.039 | 5.69 ± 0.058 |
3.531 ± 0.03 | 9.696 ± 0.11 |
1.665 ± 0.026 | 4.061 ± 0.024 |
2.478 ± 0.035 | 10.351 ± 0.056 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.174 ± 0.027 | 2.193 ± 0.041 |
5.915 ± 0.059 | 2.784 ± 0.026 |
8.037 ± 0.077 | 5.142 ± 0.061 |
4.992 ± 0.075 | 7.425 ± 0.037 |
1.411 ± 0.023 | 1.958 ± 0.023 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
