
Porcine stool-associated circular virus 7
Taxonomy: Viruses; Monodnaviria; Shotokuvirae; Cressdnaviricota; Arfiviricetes; Cremevirales; Smacoviridae; Porprismacovirus; Porprismacovirus porci7
Average proteome isoelectric point is 6.7
Get precalculated fractions of proteins
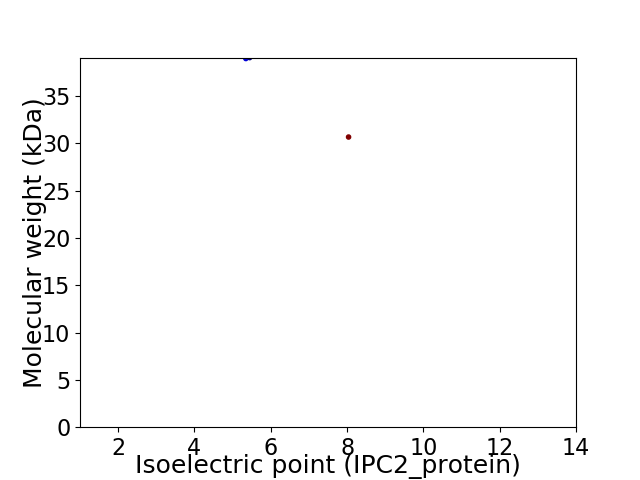
Virtual 2D-PAGE plot for 2 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A076VJL0|A0A076VJL0_9VIRU Cap protein OS=Porcine stool-associated circular virus 7 OX=1537166 PE=4 SV=1
MM1 pKa = 7.22ATNFATASYY10 pKa = 10.64QEE12 pKa = 4.43VVDD15 pKa = 4.2LHH17 pKa = 6.07TEE19 pKa = 4.0SKK21 pKa = 8.04TVSVIGIHH29 pKa = 5.77TPNTSTPVKK38 pKa = 9.44MLQGFWKK45 pKa = 10.02QFRR48 pKa = 11.84KK49 pKa = 9.98VKK51 pKa = 10.76YY52 pKa = 9.31LGCSLSLVPAARR64 pKa = 11.84LPADD68 pKa = 4.01PLQVSIGAGEE78 pKa = 3.98PTIDD82 pKa = 4.39PRR84 pKa = 11.84DD85 pKa = 3.69MLNPIMFHH93 pKa = 6.27GCHH96 pKa = 6.47GDD98 pKa = 3.58DD99 pKa = 3.94MGAILDD105 pKa = 3.85TLYY108 pKa = 11.01SGEE111 pKa = 4.22AVGTSDD117 pKa = 4.66VVRR120 pKa = 11.84KK121 pKa = 10.06QSDD124 pKa = 3.75SAVLDD129 pKa = 3.75VFSEE133 pKa = 4.18NQKK136 pKa = 10.92GNDD139 pKa = 3.7YY140 pKa = 11.28VDD142 pKa = 3.18ALYY145 pKa = 10.84YY146 pKa = 10.51RR147 pKa = 11.84SLTDD151 pKa = 3.4RR152 pKa = 11.84TWAKK156 pKa = 10.43AHH158 pKa = 5.87PQVGFRR164 pKa = 11.84KK165 pKa = 9.91SGLRR169 pKa = 11.84PLVHH173 pKa = 6.45QIVANRR179 pKa = 11.84PFSQFSPDD187 pKa = 3.3EE188 pKa = 4.07RR189 pKa = 11.84SGNVGPVVVDD199 pKa = 3.71HH200 pKa = 6.3GTSHH204 pKa = 7.5ASGQIGININNGSPTVSAVPGLPDD228 pKa = 3.33TNAVEE233 pKa = 4.39TEE235 pKa = 4.15VNFDD239 pKa = 3.07IGNGGGVGFAFATSGLRR256 pKa = 11.84PLGWQDD262 pKa = 3.34TQSSVMTSSEE272 pKa = 4.42YY273 pKa = 10.69VSEE276 pKa = 3.97PLIGNAEE283 pKa = 3.67QDD285 pKa = 3.51AQKK288 pKa = 10.67VANVWKK294 pKa = 10.42QVVVPNTIPKK304 pKa = 9.8FYY306 pKa = 9.87MGMILLPPAYY316 pKa = 8.63KK317 pKa = 9.67TEE319 pKa = 3.48QWFRR323 pKa = 11.84LQINHH328 pKa = 5.9NFAFKK333 pKa = 10.35GFRR336 pKa = 11.84GISMIDD342 pKa = 3.86DD343 pKa = 4.09DD344 pKa = 5.78PEE346 pKa = 4.46VINNAPAYY354 pKa = 9.84TNFQQ358 pKa = 3.25
MM1 pKa = 7.22ATNFATASYY10 pKa = 10.64QEE12 pKa = 4.43VVDD15 pKa = 4.2LHH17 pKa = 6.07TEE19 pKa = 4.0SKK21 pKa = 8.04TVSVIGIHH29 pKa = 5.77TPNTSTPVKK38 pKa = 9.44MLQGFWKK45 pKa = 10.02QFRR48 pKa = 11.84KK49 pKa = 9.98VKK51 pKa = 10.76YY52 pKa = 9.31LGCSLSLVPAARR64 pKa = 11.84LPADD68 pKa = 4.01PLQVSIGAGEE78 pKa = 3.98PTIDD82 pKa = 4.39PRR84 pKa = 11.84DD85 pKa = 3.69MLNPIMFHH93 pKa = 6.27GCHH96 pKa = 6.47GDD98 pKa = 3.58DD99 pKa = 3.94MGAILDD105 pKa = 3.85TLYY108 pKa = 11.01SGEE111 pKa = 4.22AVGTSDD117 pKa = 4.66VVRR120 pKa = 11.84KK121 pKa = 10.06QSDD124 pKa = 3.75SAVLDD129 pKa = 3.75VFSEE133 pKa = 4.18NQKK136 pKa = 10.92GNDD139 pKa = 3.7YY140 pKa = 11.28VDD142 pKa = 3.18ALYY145 pKa = 10.84YY146 pKa = 10.51RR147 pKa = 11.84SLTDD151 pKa = 3.4RR152 pKa = 11.84TWAKK156 pKa = 10.43AHH158 pKa = 5.87PQVGFRR164 pKa = 11.84KK165 pKa = 9.91SGLRR169 pKa = 11.84PLVHH173 pKa = 6.45QIVANRR179 pKa = 11.84PFSQFSPDD187 pKa = 3.3EE188 pKa = 4.07RR189 pKa = 11.84SGNVGPVVVDD199 pKa = 3.71HH200 pKa = 6.3GTSHH204 pKa = 7.5ASGQIGININNGSPTVSAVPGLPDD228 pKa = 3.33TNAVEE233 pKa = 4.39TEE235 pKa = 4.15VNFDD239 pKa = 3.07IGNGGGVGFAFATSGLRR256 pKa = 11.84PLGWQDD262 pKa = 3.34TQSSVMTSSEE272 pKa = 4.42YY273 pKa = 10.69VSEE276 pKa = 3.97PLIGNAEE283 pKa = 3.67QDD285 pKa = 3.51AQKK288 pKa = 10.67VANVWKK294 pKa = 10.42QVVVPNTIPKK304 pKa = 9.8FYY306 pKa = 9.87MGMILLPPAYY316 pKa = 8.63KK317 pKa = 9.67TEE319 pKa = 3.48QWFRR323 pKa = 11.84LQINHH328 pKa = 5.9NFAFKK333 pKa = 10.35GFRR336 pKa = 11.84GISMIDD342 pKa = 3.86DD343 pKa = 4.09DD344 pKa = 5.78PEE346 pKa = 4.46VINNAPAYY354 pKa = 9.84TNFQQ358 pKa = 3.25
Molecular weight: 38.95 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A076VJL0|A0A076VJL0_9VIRU Cap protein OS=Porcine stool-associated circular virus 7 OX=1537166 PE=4 SV=1
MM1 pKa = 7.32TKK3 pKa = 8.3TWMLTVPRR11 pKa = 11.84NNTAKK16 pKa = 9.95EE17 pKa = 3.75WVAISKK23 pKa = 8.8WLRR26 pKa = 11.84ANDD29 pKa = 3.33VHH31 pKa = 8.35KK32 pKa = 9.84WICAMEE38 pKa = 4.31TGADD42 pKa = 5.08GYY44 pKa = 9.42DD45 pKa = 2.85HH46 pKa = 6.3WQIRR50 pKa = 11.84LQVNKK55 pKa = 8.64TWEE58 pKa = 4.05KK59 pKa = 10.86LKK61 pKa = 10.98EE62 pKa = 3.87EE63 pKa = 4.62WGPKK67 pKa = 9.21AHH69 pKa = 6.92IEE71 pKa = 4.06EE72 pKa = 5.01ASDD75 pKa = 2.99VWDD78 pKa = 3.8YY79 pKa = 11.22EE80 pKa = 4.35RR81 pKa = 11.84KK82 pKa = 9.86SGLFFSSQDD91 pKa = 3.26TPEE94 pKa = 3.72VRR96 pKa = 11.84KK97 pKa = 10.18CRR99 pKa = 11.84FGHH102 pKa = 4.92LTWRR106 pKa = 11.84QKK108 pKa = 10.92AVLRR112 pKa = 11.84AVQSTNDD119 pKa = 3.42RR120 pKa = 11.84QVVVWYY126 pKa = 10.15DD127 pKa = 3.21PDD129 pKa = 4.09GNKK132 pKa = 10.05GKK134 pKa = 10.18SWLLGHH140 pKa = 7.53LYY142 pKa = 8.31EE143 pKa = 5.2TGQAWVVQAQDD154 pKa = 3.45TVKK157 pKa = 11.21GIIQDD162 pKa = 3.64VASEE166 pKa = 4.4YY167 pKa = 10.39INHH170 pKa = 6.35GWRR173 pKa = 11.84PMVVIDD179 pKa = 5.68IPRR182 pKa = 11.84TWRR185 pKa = 11.84WTSDD189 pKa = 3.05LYY191 pKa = 11.47VAIEE195 pKa = 4.27RR196 pKa = 11.84IKK198 pKa = 11.05DD199 pKa = 3.64GLIKK203 pKa = 10.46DD204 pKa = 3.78PRR206 pKa = 11.84YY207 pKa = 9.95SSKK210 pKa = 9.35TVHH213 pKa = 5.66IRR215 pKa = 11.84GVKK218 pKa = 9.81ILVTCNTMPKK228 pKa = 9.74LDD230 pKa = 4.44KK231 pKa = 10.84LSEE234 pKa = 4.18DD235 pKa = 2.8RR236 pKa = 11.84WIIIDD241 pKa = 3.8GDD243 pKa = 3.56ALEE246 pKa = 4.72RR247 pKa = 11.84LRR249 pKa = 11.84SAWRR253 pKa = 11.84IGEE256 pKa = 4.46GEE258 pKa = 4.05HH259 pKa = 7.23SLTT262 pKa = 4.57
MM1 pKa = 7.32TKK3 pKa = 8.3TWMLTVPRR11 pKa = 11.84NNTAKK16 pKa = 9.95EE17 pKa = 3.75WVAISKK23 pKa = 8.8WLRR26 pKa = 11.84ANDD29 pKa = 3.33VHH31 pKa = 8.35KK32 pKa = 9.84WICAMEE38 pKa = 4.31TGADD42 pKa = 5.08GYY44 pKa = 9.42DD45 pKa = 2.85HH46 pKa = 6.3WQIRR50 pKa = 11.84LQVNKK55 pKa = 8.64TWEE58 pKa = 4.05KK59 pKa = 10.86LKK61 pKa = 10.98EE62 pKa = 3.87EE63 pKa = 4.62WGPKK67 pKa = 9.21AHH69 pKa = 6.92IEE71 pKa = 4.06EE72 pKa = 5.01ASDD75 pKa = 2.99VWDD78 pKa = 3.8YY79 pKa = 11.22EE80 pKa = 4.35RR81 pKa = 11.84KK82 pKa = 9.86SGLFFSSQDD91 pKa = 3.26TPEE94 pKa = 3.72VRR96 pKa = 11.84KK97 pKa = 10.18CRR99 pKa = 11.84FGHH102 pKa = 4.92LTWRR106 pKa = 11.84QKK108 pKa = 10.92AVLRR112 pKa = 11.84AVQSTNDD119 pKa = 3.42RR120 pKa = 11.84QVVVWYY126 pKa = 10.15DD127 pKa = 3.21PDD129 pKa = 4.09GNKK132 pKa = 10.05GKK134 pKa = 10.18SWLLGHH140 pKa = 7.53LYY142 pKa = 8.31EE143 pKa = 5.2TGQAWVVQAQDD154 pKa = 3.45TVKK157 pKa = 11.21GIIQDD162 pKa = 3.64VASEE166 pKa = 4.4YY167 pKa = 10.39INHH170 pKa = 6.35GWRR173 pKa = 11.84PMVVIDD179 pKa = 5.68IPRR182 pKa = 11.84TWRR185 pKa = 11.84WTSDD189 pKa = 3.05LYY191 pKa = 11.47VAIEE195 pKa = 4.27RR196 pKa = 11.84IKK198 pKa = 11.05DD199 pKa = 3.64GLIKK203 pKa = 10.46DD204 pKa = 3.78PRR206 pKa = 11.84YY207 pKa = 9.95SSKK210 pKa = 9.35TVHH213 pKa = 5.66IRR215 pKa = 11.84GVKK218 pKa = 9.81ILVTCNTMPKK228 pKa = 9.74LDD230 pKa = 4.44KK231 pKa = 10.84LSEE234 pKa = 4.18DD235 pKa = 2.8RR236 pKa = 11.84WIIIDD241 pKa = 3.8GDD243 pKa = 3.56ALEE246 pKa = 4.72RR247 pKa = 11.84LRR249 pKa = 11.84SAWRR253 pKa = 11.84IGEE256 pKa = 4.46GEE258 pKa = 4.05HH259 pKa = 7.23SLTT262 pKa = 4.57
Molecular weight: 30.66 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
620 |
262 |
358 |
310.0 |
34.8 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.613 ± 0.545 | 0.806 ± 0.208 |
6.613 ± 0.392 | 4.677 ± 0.877 |
3.226 ± 1.277 | 7.581 ± 0.904 |
2.742 ± 0.191 | 5.806 ± 0.653 |
5.484 ± 1.319 | 6.452 ± 0.257 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.258 ± 0.215 | 4.677 ± 0.996 |
5.161 ± 1.293 | 4.516 ± 0.429 |
5.161 ± 1.283 | 6.774 ± 0.878 |
6.29 ± 0.122 | 8.871 ± 0.525 |
3.548 ± 1.804 | 2.742 ± 0.043 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
