
Eidolon helvum parvovirus 1 isolate Bat/Ghana/-/2009
Taxonomy: Viruses; Monodnaviria; Shotokuvirae; Cossaviricota; Quintoviricetes; Piccovirales; Parvoviridae; Parvovirinae; Tetraparvovirus; Chiropteran tetraparvovirus 1
Average proteome isoelectric point is 5.96
Get precalculated fractions of proteins
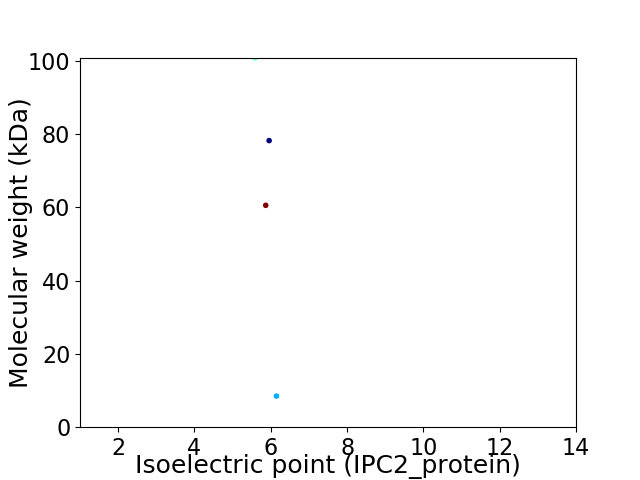
Virtual 2D-PAGE plot for 4 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|H2ESE6|H2ESE6_BTPV1 MARF OS=Eidolon helvum parvovirus 1 (isolate Bat/Ghana/-/2009) OX=1560037 GN=MARF PE=4 SV=1
MM1 pKa = 7.71ASGGVAFFMPTGEE14 pKa = 4.14GGKK17 pKa = 10.18AISKK21 pKa = 10.25LYY23 pKa = 10.79DD24 pKa = 3.41GAVTGWSQSHH34 pKa = 6.4LRR36 pKa = 11.84HH37 pKa = 4.44GHH39 pKa = 4.75YY40 pKa = 10.14RR41 pKa = 11.84ATALSGLVYY50 pKa = 10.46QKK52 pKa = 11.23AFEE55 pKa = 4.33NLDD58 pKa = 3.42AFAKK62 pKa = 10.14HH63 pKa = 6.19LPPSLRR69 pKa = 11.84LVQTLYY75 pKa = 11.59NNISKK80 pKa = 9.65QRR82 pKa = 11.84NKK84 pKa = 9.17TANTEE89 pKa = 3.88WWWRR93 pKa = 11.84VYY95 pKa = 10.0TDD97 pKa = 3.54CTRR100 pKa = 11.84LLLTVAQPQAADD112 pKa = 3.38ALRR115 pKa = 11.84RR116 pKa = 11.84AWNGTSPLRR125 pKa = 11.84PPAQQVLQNVYY136 pKa = 7.93MLAYY140 pKa = 9.56NSHH143 pKa = 6.34IPSEE147 pKa = 4.13EE148 pKa = 3.95DD149 pKa = 3.11LNTFFDD155 pKa = 4.02EE156 pKa = 4.49SVLGSVSGDD165 pKa = 3.3PEE167 pKa = 3.52NFAFVKK173 pKa = 9.94QAVVSAFHH181 pKa = 6.79PADD184 pKa = 3.67GGAATEE190 pKa = 4.1QRR192 pKa = 11.84SDD194 pKa = 3.66VPEE197 pKa = 4.49SPSPPSEE204 pKa = 4.1SDD206 pKa = 2.75HH207 pKa = 6.05SQPSGGGLLVPGYY220 pKa = 10.84NYY222 pKa = 10.28VGPGNPLDD230 pKa = 4.14NGPPKK235 pKa = 10.92GPVDD239 pKa = 3.48EE240 pKa = 4.81AARR243 pKa = 11.84NHH245 pKa = 5.92DD246 pKa = 3.31RR247 pKa = 11.84RR248 pKa = 11.84YY249 pKa = 11.27DD250 pKa = 3.49EE251 pKa = 4.53MLSHH255 pKa = 7.15GDD257 pKa = 3.43VPYY260 pKa = 10.98LDD262 pKa = 4.03SQGVDD267 pKa = 2.97QLMTKK272 pKa = 10.17EE273 pKa = 4.24IEE275 pKa = 4.22DD276 pKa = 3.94AEE278 pKa = 4.48KK279 pKa = 10.75KK280 pKa = 10.46EE281 pKa = 3.93PLGPIDD287 pKa = 4.78ALVANAARR295 pKa = 11.84ALWKK299 pKa = 10.64GKK301 pKa = 7.71EE302 pKa = 4.11TLASVIGDD310 pKa = 3.27QGHH313 pKa = 5.49QVLPPNPPSVDD324 pKa = 3.27QGSHH328 pKa = 6.27LGSKK332 pKa = 10.35RR333 pKa = 11.84PLEE336 pKa = 4.11EE337 pKa = 4.16SEE339 pKa = 4.56EE340 pKa = 4.3NSDD343 pKa = 3.94SLAKK347 pKa = 10.32KK348 pKa = 9.46FRR350 pKa = 11.84PGSSPKK356 pKa = 10.12SDD358 pKa = 4.1LEE360 pKa = 4.23PSGGTAPTAPPKK372 pKa = 10.19GATMSTEE379 pKa = 4.11AEE381 pKa = 4.29GSGGGIKK388 pKa = 10.19VKK390 pKa = 10.67ALWTGGTEE398 pKa = 4.18FSDD401 pKa = 3.78TSIHH405 pKa = 5.85TSHH408 pKa = 6.51TRR410 pKa = 11.84VALLADD416 pKa = 4.48RR417 pKa = 11.84GTYY420 pKa = 8.58MPIYY424 pKa = 10.12RR425 pKa = 11.84PGEE428 pKa = 4.2TTSVIQPLLGMVTPYY443 pKa = 10.98SYY445 pKa = 10.96IDD447 pKa = 3.74VNSLSAHH454 pKa = 5.52LTPRR458 pKa = 11.84DD459 pKa = 3.66FQQLIDD465 pKa = 4.08EE466 pKa = 4.57YY467 pKa = 11.71GEE469 pKa = 3.95IRR471 pKa = 11.84PKK473 pKa = 10.79SLTIGISGIVVKK485 pKa = 10.53DD486 pKa = 3.25VSVNTTGTAVTDD498 pKa = 3.77SGSGGITVFADD509 pKa = 2.94EE510 pKa = 5.56GYY512 pKa = 10.61DD513 pKa = 3.5YY514 pKa = 10.84PYY516 pKa = 11.53VLGHH520 pKa = 6.2NQDD523 pKa = 3.85TLPGHH528 pKa = 7.2LPGEE532 pKa = 4.69HH533 pKa = 6.1YY534 pKa = 11.06VLPQYY539 pKa = 10.85AYY541 pKa = 7.83CTRR544 pKa = 11.84GRR546 pKa = 11.84EE547 pKa = 3.9VVNANKK553 pKa = 9.73IDD555 pKa = 4.13IIQDD559 pKa = 2.9HH560 pKa = 5.95RR561 pKa = 11.84TDD563 pKa = 4.11FFLLEE568 pKa = 4.02HH569 pKa = 7.08HH570 pKa = 7.43DD571 pKa = 3.95AXCLNSGDD579 pKa = 4.23TWSHH583 pKa = 6.09TYY585 pKa = 10.67SFPDD589 pKa = 3.35LPFRR593 pKa = 11.84RR594 pKa = 11.84LTYY597 pKa = 10.04PSQHH601 pKa = 6.79LYY603 pKa = 9.59AQHH606 pKa = 6.06NPQMKK611 pKa = 9.74SRR613 pKa = 11.84LGVFRR618 pKa = 11.84SVDD621 pKa = 3.66QNGDD625 pKa = 3.78PAWQRR630 pKa = 11.84LRR632 pKa = 11.84GSDD635 pKa = 4.25IGQLPCNYY643 pKa = 10.11LPGPQANLPTTSDD656 pKa = 3.75FQKK659 pKa = 10.72GDD661 pKa = 3.24SMLPAAIGDD670 pKa = 4.72PITGDD675 pKa = 3.25RR676 pKa = 11.84YY677 pKa = 8.09TVTPLVHH684 pKa = 6.16QPWSMVHH691 pKa = 7.23DD692 pKa = 4.96EE693 pKa = 4.5YY694 pKa = 11.76NNTGQRR700 pKa = 11.84TQSVVSTVGSVAYY713 pKa = 8.96MPRR716 pKa = 11.84KK717 pKa = 9.77HH718 pKa = 6.21EE719 pKa = 4.07EE720 pKa = 4.34SYY722 pKa = 11.4GSPNDD727 pKa = 3.6ALTEE731 pKa = 3.79ITTPMRR737 pKa = 11.84VVQEE741 pKa = 3.99QADD744 pKa = 4.23LVSSRR749 pKa = 11.84LGHH752 pKa = 5.85TFMIPSFSRR761 pKa = 11.84KK762 pKa = 9.45AGSASTTTYY771 pKa = 10.51NEE773 pKa = 3.73PMFPLLPGAVWNPNPLTYY791 pKa = 10.47DD792 pKa = 3.39CQIWCKK798 pKa = 10.76VPDD801 pKa = 4.11TEE803 pKa = 5.14CKK805 pKa = 10.51FMTQYY810 pKa = 11.1PLLGGWGMDD819 pKa = 3.83TPPPMIFLRR828 pKa = 11.84IRR830 pKa = 11.84RR831 pKa = 11.84QPGPPGPGAHH841 pKa = 5.63TVPGSTLNQYY851 pKa = 10.73VIFHH855 pKa = 6.48LHH857 pKa = 5.43YY858 pKa = 11.1SMEE861 pKa = 4.22FEE863 pKa = 3.9VRR865 pKa = 11.84RR866 pKa = 11.84RR867 pKa = 11.84KK868 pKa = 9.8RR869 pKa = 11.84SRR871 pKa = 11.84RR872 pKa = 11.84HH873 pKa = 4.96NPEE876 pKa = 3.3MPAPFPQTWSGKK888 pKa = 8.12MPYY891 pKa = 9.11TLSTDD896 pKa = 3.29GKK898 pKa = 10.55SGPEE902 pKa = 3.72YY903 pKa = 10.54NVPRR907 pKa = 11.84DD908 pKa = 3.17QWVARR913 pKa = 11.84NYY915 pKa = 10.72SQLLL919 pKa = 3.6
MM1 pKa = 7.71ASGGVAFFMPTGEE14 pKa = 4.14GGKK17 pKa = 10.18AISKK21 pKa = 10.25LYY23 pKa = 10.79DD24 pKa = 3.41GAVTGWSQSHH34 pKa = 6.4LRR36 pKa = 11.84HH37 pKa = 4.44GHH39 pKa = 4.75YY40 pKa = 10.14RR41 pKa = 11.84ATALSGLVYY50 pKa = 10.46QKK52 pKa = 11.23AFEE55 pKa = 4.33NLDD58 pKa = 3.42AFAKK62 pKa = 10.14HH63 pKa = 6.19LPPSLRR69 pKa = 11.84LVQTLYY75 pKa = 11.59NNISKK80 pKa = 9.65QRR82 pKa = 11.84NKK84 pKa = 9.17TANTEE89 pKa = 3.88WWWRR93 pKa = 11.84VYY95 pKa = 10.0TDD97 pKa = 3.54CTRR100 pKa = 11.84LLLTVAQPQAADD112 pKa = 3.38ALRR115 pKa = 11.84RR116 pKa = 11.84AWNGTSPLRR125 pKa = 11.84PPAQQVLQNVYY136 pKa = 7.93MLAYY140 pKa = 9.56NSHH143 pKa = 6.34IPSEE147 pKa = 4.13EE148 pKa = 3.95DD149 pKa = 3.11LNTFFDD155 pKa = 4.02EE156 pKa = 4.49SVLGSVSGDD165 pKa = 3.3PEE167 pKa = 3.52NFAFVKK173 pKa = 9.94QAVVSAFHH181 pKa = 6.79PADD184 pKa = 3.67GGAATEE190 pKa = 4.1QRR192 pKa = 11.84SDD194 pKa = 3.66VPEE197 pKa = 4.49SPSPPSEE204 pKa = 4.1SDD206 pKa = 2.75HH207 pKa = 6.05SQPSGGGLLVPGYY220 pKa = 10.84NYY222 pKa = 10.28VGPGNPLDD230 pKa = 4.14NGPPKK235 pKa = 10.92GPVDD239 pKa = 3.48EE240 pKa = 4.81AARR243 pKa = 11.84NHH245 pKa = 5.92DD246 pKa = 3.31RR247 pKa = 11.84RR248 pKa = 11.84YY249 pKa = 11.27DD250 pKa = 3.49EE251 pKa = 4.53MLSHH255 pKa = 7.15GDD257 pKa = 3.43VPYY260 pKa = 10.98LDD262 pKa = 4.03SQGVDD267 pKa = 2.97QLMTKK272 pKa = 10.17EE273 pKa = 4.24IEE275 pKa = 4.22DD276 pKa = 3.94AEE278 pKa = 4.48KK279 pKa = 10.75KK280 pKa = 10.46EE281 pKa = 3.93PLGPIDD287 pKa = 4.78ALVANAARR295 pKa = 11.84ALWKK299 pKa = 10.64GKK301 pKa = 7.71EE302 pKa = 4.11TLASVIGDD310 pKa = 3.27QGHH313 pKa = 5.49QVLPPNPPSVDD324 pKa = 3.27QGSHH328 pKa = 6.27LGSKK332 pKa = 10.35RR333 pKa = 11.84PLEE336 pKa = 4.11EE337 pKa = 4.16SEE339 pKa = 4.56EE340 pKa = 4.3NSDD343 pKa = 3.94SLAKK347 pKa = 10.32KK348 pKa = 9.46FRR350 pKa = 11.84PGSSPKK356 pKa = 10.12SDD358 pKa = 4.1LEE360 pKa = 4.23PSGGTAPTAPPKK372 pKa = 10.19GATMSTEE379 pKa = 4.11AEE381 pKa = 4.29GSGGGIKK388 pKa = 10.19VKK390 pKa = 10.67ALWTGGTEE398 pKa = 4.18FSDD401 pKa = 3.78TSIHH405 pKa = 5.85TSHH408 pKa = 6.51TRR410 pKa = 11.84VALLADD416 pKa = 4.48RR417 pKa = 11.84GTYY420 pKa = 8.58MPIYY424 pKa = 10.12RR425 pKa = 11.84PGEE428 pKa = 4.2TTSVIQPLLGMVTPYY443 pKa = 10.98SYY445 pKa = 10.96IDD447 pKa = 3.74VNSLSAHH454 pKa = 5.52LTPRR458 pKa = 11.84DD459 pKa = 3.66FQQLIDD465 pKa = 4.08EE466 pKa = 4.57YY467 pKa = 11.71GEE469 pKa = 3.95IRR471 pKa = 11.84PKK473 pKa = 10.79SLTIGISGIVVKK485 pKa = 10.53DD486 pKa = 3.25VSVNTTGTAVTDD498 pKa = 3.77SGSGGITVFADD509 pKa = 2.94EE510 pKa = 5.56GYY512 pKa = 10.61DD513 pKa = 3.5YY514 pKa = 10.84PYY516 pKa = 11.53VLGHH520 pKa = 6.2NQDD523 pKa = 3.85TLPGHH528 pKa = 7.2LPGEE532 pKa = 4.69HH533 pKa = 6.1YY534 pKa = 11.06VLPQYY539 pKa = 10.85AYY541 pKa = 7.83CTRR544 pKa = 11.84GRR546 pKa = 11.84EE547 pKa = 3.9VVNANKK553 pKa = 9.73IDD555 pKa = 4.13IIQDD559 pKa = 2.9HH560 pKa = 5.95RR561 pKa = 11.84TDD563 pKa = 4.11FFLLEE568 pKa = 4.02HH569 pKa = 7.08HH570 pKa = 7.43DD571 pKa = 3.95AXCLNSGDD579 pKa = 4.23TWSHH583 pKa = 6.09TYY585 pKa = 10.67SFPDD589 pKa = 3.35LPFRR593 pKa = 11.84RR594 pKa = 11.84LTYY597 pKa = 10.04PSQHH601 pKa = 6.79LYY603 pKa = 9.59AQHH606 pKa = 6.06NPQMKK611 pKa = 9.74SRR613 pKa = 11.84LGVFRR618 pKa = 11.84SVDD621 pKa = 3.66QNGDD625 pKa = 3.78PAWQRR630 pKa = 11.84LRR632 pKa = 11.84GSDD635 pKa = 4.25IGQLPCNYY643 pKa = 10.11LPGPQANLPTTSDD656 pKa = 3.75FQKK659 pKa = 10.72GDD661 pKa = 3.24SMLPAAIGDD670 pKa = 4.72PITGDD675 pKa = 3.25RR676 pKa = 11.84YY677 pKa = 8.09TVTPLVHH684 pKa = 6.16QPWSMVHH691 pKa = 7.23DD692 pKa = 4.96EE693 pKa = 4.5YY694 pKa = 11.76NNTGQRR700 pKa = 11.84TQSVVSTVGSVAYY713 pKa = 8.96MPRR716 pKa = 11.84KK717 pKa = 9.77HH718 pKa = 6.21EE719 pKa = 4.07EE720 pKa = 4.34SYY722 pKa = 11.4GSPNDD727 pKa = 3.6ALTEE731 pKa = 3.79ITTPMRR737 pKa = 11.84VVQEE741 pKa = 3.99QADD744 pKa = 4.23LVSSRR749 pKa = 11.84LGHH752 pKa = 5.85TFMIPSFSRR761 pKa = 11.84KK762 pKa = 9.45AGSASTTTYY771 pKa = 10.51NEE773 pKa = 3.73PMFPLLPGAVWNPNPLTYY791 pKa = 10.47DD792 pKa = 3.39CQIWCKK798 pKa = 10.76VPDD801 pKa = 4.11TEE803 pKa = 5.14CKK805 pKa = 10.51FMTQYY810 pKa = 11.1PLLGGWGMDD819 pKa = 3.83TPPPMIFLRR828 pKa = 11.84IRR830 pKa = 11.84RR831 pKa = 11.84QPGPPGPGAHH841 pKa = 5.63TVPGSTLNQYY851 pKa = 10.73VIFHH855 pKa = 6.48LHH857 pKa = 5.43YY858 pKa = 11.1SMEE861 pKa = 4.22FEE863 pKa = 3.9VRR865 pKa = 11.84RR866 pKa = 11.84RR867 pKa = 11.84KK868 pKa = 9.8RR869 pKa = 11.84SRR871 pKa = 11.84RR872 pKa = 11.84HH873 pKa = 4.96NPEE876 pKa = 3.3MPAPFPQTWSGKK888 pKa = 8.12MPYY891 pKa = 9.11TLSTDD896 pKa = 3.29GKK898 pKa = 10.55SGPEE902 pKa = 3.72YY903 pKa = 10.54NVPRR907 pKa = 11.84DD908 pKa = 3.17QWVARR913 pKa = 11.84NYY915 pKa = 10.72SQLLL919 pKa = 3.6
Molecular weight: 100.92 kDa
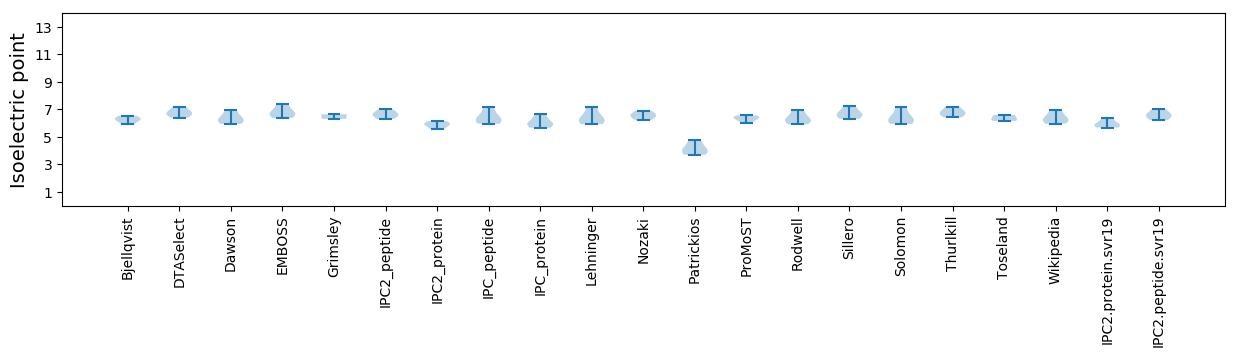
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|H2ESE7|H2ESE7_BTPV1 Putative capsid protein VP2 OS=Eidolon helvum parvovirus 1 (isolate Bat/Ghana/-/2009) OX=1560037 PE=3 SV=1
MM1 pKa = 7.86EE2 pKa = 5.18EE3 pKa = 4.31PLLNSALTSLKK14 pKa = 10.54VLAPLVNQTIHH25 pKa = 6.54SLAEE29 pKa = 4.15VVSLSPATIMWVLVILWIMAPLRR52 pKa = 11.84AQWMRR57 pKa = 11.84QLAIMTEE64 pKa = 4.07GMTKK68 pKa = 10.31CFPTGTSPTT77 pKa = 3.75
MM1 pKa = 7.86EE2 pKa = 5.18EE3 pKa = 4.31PLLNSALTSLKK14 pKa = 10.54VLAPLVNQTIHH25 pKa = 6.54SLAEE29 pKa = 4.15VVSLSPATIMWVLVILWIMAPLRR52 pKa = 11.84AQWMRR57 pKa = 11.84QLAIMTEE64 pKa = 4.07GMTKK68 pKa = 10.31CFPTGTSPTT77 pKa = 3.75
Molecular weight: 8.49 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
2232 |
77 |
919 |
558.0 |
62.07 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.944 ± 1.141 | 1.344 ± 0.407 |
5.421 ± 0.469 | 5.242 ± 0.813 |
3.226 ± 0.3 | 7.93 ± 0.603 |
2.912 ± 0.475 | 3.584 ± 0.403 |
3.405 ± 0.37 | 8.244 ± 0.6 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.419 ± 0.583 | 3.539 ± 0.303 |
8.065 ± 0.9 | 4.077 ± 0.564 |
5.914 ± 0.68 | 7.706 ± 0.337 |
6.765 ± 1.054 | 6.676 ± 0.375 |
2.509 ± 0.729 | 3.987 ± 0.426 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
