
Adeno-associated virus 2 (isolate Srivastava/1982) (AAV-2)
Taxonomy: Viruses; Monodnaviria; Shotokuvirae; Cossaviricota; Quintoviricetes; Piccovirales; Parvoviridae; Parvovirinae; Dependoparvovirus; Adeno-associated dependoparvovirus A; Adeno-associated virus - 2
Average proteome isoelectric point is 6.56
Get precalculated fractions of proteins
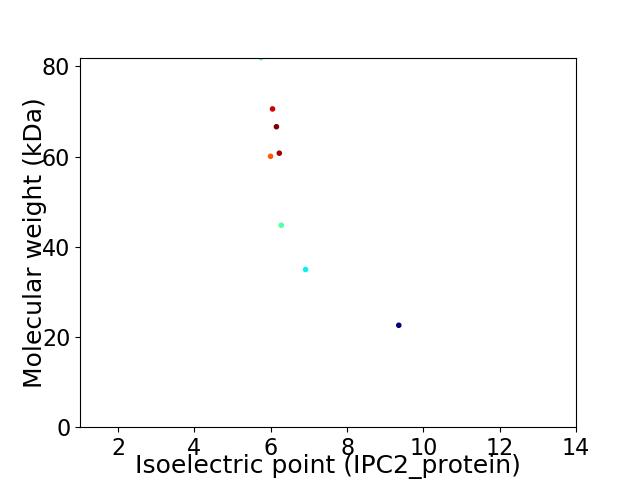
Virtual 2D-PAGE plot for 8 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>sp|Q89268|REP78_AAV2S Protein Rep78 OS=Adeno-associated virus 2 (isolate Srivastava/1982) OX=648242 GN=Rep78 PE=1 SV=1
MM1 pKa = 7.47AADD4 pKa = 5.48GYY6 pKa = 11.26LPDD9 pKa = 3.96WLEE12 pKa = 4.24DD13 pKa = 3.65TLSEE17 pKa = 6.15GIRR20 pKa = 11.84QWWKK24 pKa = 10.59LKK26 pKa = 10.21PGPPPPKK33 pKa = 8.73PAEE36 pKa = 3.89RR37 pKa = 11.84HH38 pKa = 5.74KK39 pKa = 11.12DD40 pKa = 3.41DD41 pKa = 4.29SRR43 pKa = 11.84GLVLPGYY50 pKa = 10.19KK51 pKa = 10.22YY52 pKa = 10.62LGPFNGLDD60 pKa = 2.91KK61 pKa = 11.47GEE63 pKa = 4.47PVNEE67 pKa = 3.78ADD69 pKa = 4.62AAALEE74 pKa = 4.24HH75 pKa = 7.12DD76 pKa = 3.9KK77 pKa = 11.32AYY79 pKa = 10.75DD80 pKa = 3.58RR81 pKa = 11.84QLDD84 pKa = 3.91SGDD87 pKa = 3.77NPYY90 pKa = 11.12LKK92 pKa = 10.54YY93 pKa = 10.86NHH95 pKa = 7.21ADD97 pKa = 3.17AEE99 pKa = 4.31FQEE102 pKa = 4.46RR103 pKa = 11.84LKK105 pKa = 11.03EE106 pKa = 3.98DD107 pKa = 3.3TSFGGNLGRR116 pKa = 11.84AVFQAKK122 pKa = 9.74KK123 pKa = 10.24RR124 pKa = 11.84VLEE127 pKa = 3.81PLGLVEE133 pKa = 5.13EE134 pKa = 4.6PVKK137 pKa = 9.85TAPGKK142 pKa = 9.96KK143 pKa = 9.36RR144 pKa = 11.84PVEE147 pKa = 3.98HH148 pKa = 6.89SPVEE152 pKa = 4.15PDD154 pKa = 3.16SSSGTGKK161 pKa = 10.46AGQQPARR168 pKa = 11.84KK169 pKa = 8.94RR170 pKa = 11.84LNFGQTGDD178 pKa = 3.98ADD180 pKa = 4.25SVPDD184 pKa = 4.6PQPLGQPPAAPSGLGTNTMATGSGAPMADD213 pKa = 3.37NNEE216 pKa = 4.16GADD219 pKa = 3.78GVGNSSGNWHH229 pKa = 7.23CDD231 pKa = 3.35STWMGDD237 pKa = 3.49RR238 pKa = 11.84VITTSTRR245 pKa = 11.84TWALPTYY252 pKa = 10.3NNHH255 pKa = 6.8LYY257 pKa = 10.25KK258 pKa = 10.6QISSQSGASNDD269 pKa = 3.08NHH271 pKa = 6.16YY272 pKa = 10.71FGYY275 pKa = 7.95STPWGYY281 pKa = 11.2FDD283 pKa = 5.8FNRR286 pKa = 11.84FHH288 pKa = 6.91CHH290 pKa = 5.57FSPRR294 pKa = 11.84DD295 pKa = 3.43WQRR298 pKa = 11.84LINNNWGFRR307 pKa = 11.84PKK309 pKa = 10.42RR310 pKa = 11.84LNFKK314 pKa = 10.31LFNIQVKK321 pKa = 9.66EE322 pKa = 4.06VTQNDD327 pKa = 4.03GTTTIANNLTSTVQVFTDD345 pKa = 3.48SEE347 pKa = 4.49YY348 pKa = 10.74QLPYY352 pKa = 10.78VLGSAHH358 pKa = 6.25QGCLPPFPADD368 pKa = 2.94VFMVPQYY375 pKa = 11.56GYY377 pKa = 8.93LTLNNGSQAVGRR389 pKa = 11.84SSFYY393 pKa = 10.4CLEE396 pKa = 4.08YY397 pKa = 10.65FPSQMLRR404 pKa = 11.84TGNNFTFSYY413 pKa = 9.76TFEE416 pKa = 4.62DD417 pKa = 4.48VPFHH421 pKa = 6.78SSYY424 pKa = 11.56AHH426 pKa = 5.63SQSLDD431 pKa = 2.94RR432 pKa = 11.84LMNPLIDD439 pKa = 3.29QYY441 pKa = 11.66LYY443 pKa = 10.92YY444 pKa = 10.81LSRR447 pKa = 11.84TNTPSGTTTQSRR459 pKa = 11.84LQFSQAGASDD469 pKa = 3.3IRR471 pKa = 11.84DD472 pKa = 3.38QSRR475 pKa = 11.84NWLPGPCYY483 pKa = 9.9RR484 pKa = 11.84QQRR487 pKa = 11.84VSKK490 pKa = 9.9TSADD494 pKa = 3.88NNNSEE499 pKa = 4.28YY500 pKa = 10.88SWTGATKK507 pKa = 9.7YY508 pKa = 10.23HH509 pKa = 6.45LNGRR513 pKa = 11.84DD514 pKa = 3.41SLVNPGPAMASHH526 pKa = 7.23KK527 pKa = 10.69DD528 pKa = 3.53DD529 pKa = 3.69EE530 pKa = 5.0EE531 pKa = 4.99KK532 pKa = 10.71FFPQSGVLIFGKK544 pKa = 10.26QGSEE548 pKa = 3.75KK549 pKa = 10.33TNVDD553 pKa = 3.06IEE555 pKa = 4.42KK556 pKa = 10.88VMITDD561 pKa = 3.44EE562 pKa = 4.28EE563 pKa = 4.63EE564 pKa = 3.81IRR566 pKa = 11.84TTNPVATEE574 pKa = 4.08QYY576 pKa = 11.02GSVSTNLQRR585 pKa = 11.84GNRR588 pKa = 11.84QAATADD594 pKa = 3.69VNTQGVLPGMVWQDD608 pKa = 2.54RR609 pKa = 11.84DD610 pKa = 3.62VYY612 pKa = 10.93LQGPIWAKK620 pKa = 10.14IPHH623 pKa = 6.57TDD625 pKa = 3.17GHH627 pKa = 5.96FHH629 pKa = 7.43PSPLMGGFGLKK640 pKa = 10.12HH641 pKa = 6.61PPPQILIKK649 pKa = 9.58NTPVPANPSTTFSAAKK665 pKa = 8.7FASFITQYY673 pKa = 9.97STGQVSVEE681 pKa = 4.4IEE683 pKa = 3.74WEE685 pKa = 4.1LQKK688 pKa = 11.15EE689 pKa = 4.16NSKK692 pKa = 10.25RR693 pKa = 11.84WNPEE697 pKa = 2.74IQYY700 pKa = 9.56TSNYY704 pKa = 8.24NKK706 pKa = 10.29SVNVDD711 pKa = 3.7FTVDD715 pKa = 3.26TNGVYY720 pKa = 10.21SEE722 pKa = 4.47PRR724 pKa = 11.84PIGTRR729 pKa = 11.84YY730 pKa = 8.08LTRR733 pKa = 11.84NLL735 pKa = 3.55
MM1 pKa = 7.47AADD4 pKa = 5.48GYY6 pKa = 11.26LPDD9 pKa = 3.96WLEE12 pKa = 4.24DD13 pKa = 3.65TLSEE17 pKa = 6.15GIRR20 pKa = 11.84QWWKK24 pKa = 10.59LKK26 pKa = 10.21PGPPPPKK33 pKa = 8.73PAEE36 pKa = 3.89RR37 pKa = 11.84HH38 pKa = 5.74KK39 pKa = 11.12DD40 pKa = 3.41DD41 pKa = 4.29SRR43 pKa = 11.84GLVLPGYY50 pKa = 10.19KK51 pKa = 10.22YY52 pKa = 10.62LGPFNGLDD60 pKa = 2.91KK61 pKa = 11.47GEE63 pKa = 4.47PVNEE67 pKa = 3.78ADD69 pKa = 4.62AAALEE74 pKa = 4.24HH75 pKa = 7.12DD76 pKa = 3.9KK77 pKa = 11.32AYY79 pKa = 10.75DD80 pKa = 3.58RR81 pKa = 11.84QLDD84 pKa = 3.91SGDD87 pKa = 3.77NPYY90 pKa = 11.12LKK92 pKa = 10.54YY93 pKa = 10.86NHH95 pKa = 7.21ADD97 pKa = 3.17AEE99 pKa = 4.31FQEE102 pKa = 4.46RR103 pKa = 11.84LKK105 pKa = 11.03EE106 pKa = 3.98DD107 pKa = 3.3TSFGGNLGRR116 pKa = 11.84AVFQAKK122 pKa = 9.74KK123 pKa = 10.24RR124 pKa = 11.84VLEE127 pKa = 3.81PLGLVEE133 pKa = 5.13EE134 pKa = 4.6PVKK137 pKa = 9.85TAPGKK142 pKa = 9.96KK143 pKa = 9.36RR144 pKa = 11.84PVEE147 pKa = 3.98HH148 pKa = 6.89SPVEE152 pKa = 4.15PDD154 pKa = 3.16SSSGTGKK161 pKa = 10.46AGQQPARR168 pKa = 11.84KK169 pKa = 8.94RR170 pKa = 11.84LNFGQTGDD178 pKa = 3.98ADD180 pKa = 4.25SVPDD184 pKa = 4.6PQPLGQPPAAPSGLGTNTMATGSGAPMADD213 pKa = 3.37NNEE216 pKa = 4.16GADD219 pKa = 3.78GVGNSSGNWHH229 pKa = 7.23CDD231 pKa = 3.35STWMGDD237 pKa = 3.49RR238 pKa = 11.84VITTSTRR245 pKa = 11.84TWALPTYY252 pKa = 10.3NNHH255 pKa = 6.8LYY257 pKa = 10.25KK258 pKa = 10.6QISSQSGASNDD269 pKa = 3.08NHH271 pKa = 6.16YY272 pKa = 10.71FGYY275 pKa = 7.95STPWGYY281 pKa = 11.2FDD283 pKa = 5.8FNRR286 pKa = 11.84FHH288 pKa = 6.91CHH290 pKa = 5.57FSPRR294 pKa = 11.84DD295 pKa = 3.43WQRR298 pKa = 11.84LINNNWGFRR307 pKa = 11.84PKK309 pKa = 10.42RR310 pKa = 11.84LNFKK314 pKa = 10.31LFNIQVKK321 pKa = 9.66EE322 pKa = 4.06VTQNDD327 pKa = 4.03GTTTIANNLTSTVQVFTDD345 pKa = 3.48SEE347 pKa = 4.49YY348 pKa = 10.74QLPYY352 pKa = 10.78VLGSAHH358 pKa = 6.25QGCLPPFPADD368 pKa = 2.94VFMVPQYY375 pKa = 11.56GYY377 pKa = 8.93LTLNNGSQAVGRR389 pKa = 11.84SSFYY393 pKa = 10.4CLEE396 pKa = 4.08YY397 pKa = 10.65FPSQMLRR404 pKa = 11.84TGNNFTFSYY413 pKa = 9.76TFEE416 pKa = 4.62DD417 pKa = 4.48VPFHH421 pKa = 6.78SSYY424 pKa = 11.56AHH426 pKa = 5.63SQSLDD431 pKa = 2.94RR432 pKa = 11.84LMNPLIDD439 pKa = 3.29QYY441 pKa = 11.66LYY443 pKa = 10.92YY444 pKa = 10.81LSRR447 pKa = 11.84TNTPSGTTTQSRR459 pKa = 11.84LQFSQAGASDD469 pKa = 3.3IRR471 pKa = 11.84DD472 pKa = 3.38QSRR475 pKa = 11.84NWLPGPCYY483 pKa = 9.9RR484 pKa = 11.84QQRR487 pKa = 11.84VSKK490 pKa = 9.9TSADD494 pKa = 3.88NNNSEE499 pKa = 4.28YY500 pKa = 10.88SWTGATKK507 pKa = 9.7YY508 pKa = 10.23HH509 pKa = 6.45LNGRR513 pKa = 11.84DD514 pKa = 3.41SLVNPGPAMASHH526 pKa = 7.23KK527 pKa = 10.69DD528 pKa = 3.53DD529 pKa = 3.69EE530 pKa = 5.0EE531 pKa = 4.99KK532 pKa = 10.71FFPQSGVLIFGKK544 pKa = 10.26QGSEE548 pKa = 3.75KK549 pKa = 10.33TNVDD553 pKa = 3.06IEE555 pKa = 4.42KK556 pKa = 10.88VMITDD561 pKa = 3.44EE562 pKa = 4.28EE563 pKa = 4.63EE564 pKa = 3.81IRR566 pKa = 11.84TTNPVATEE574 pKa = 4.08QYY576 pKa = 11.02GSVSTNLQRR585 pKa = 11.84GNRR588 pKa = 11.84QAATADD594 pKa = 3.69VNTQGVLPGMVWQDD608 pKa = 2.54RR609 pKa = 11.84DD610 pKa = 3.62VYY612 pKa = 10.93LQGPIWAKK620 pKa = 10.14IPHH623 pKa = 6.57TDD625 pKa = 3.17GHH627 pKa = 5.96FHH629 pKa = 7.43PSPLMGGFGLKK640 pKa = 10.12HH641 pKa = 6.61PPPQILIKK649 pKa = 9.58NTPVPANPSTTFSAAKK665 pKa = 8.7FASFITQYY673 pKa = 9.97STGQVSVEE681 pKa = 4.4IEE683 pKa = 3.74WEE685 pKa = 4.1LQKK688 pKa = 11.15EE689 pKa = 4.16NSKK692 pKa = 10.25RR693 pKa = 11.84WNPEE697 pKa = 2.74IQYY700 pKa = 9.56TSNYY704 pKa = 8.24NKK706 pKa = 10.29SVNVDD711 pKa = 3.7FTVDD715 pKa = 3.26TNGVYY720 pKa = 10.21SEE722 pKa = 4.47PRR724 pKa = 11.84PIGTRR729 pKa = 11.84YY730 pKa = 8.08LTRR733 pKa = 11.84NLL735 pKa = 3.55
Molecular weight: 81.94 kDa
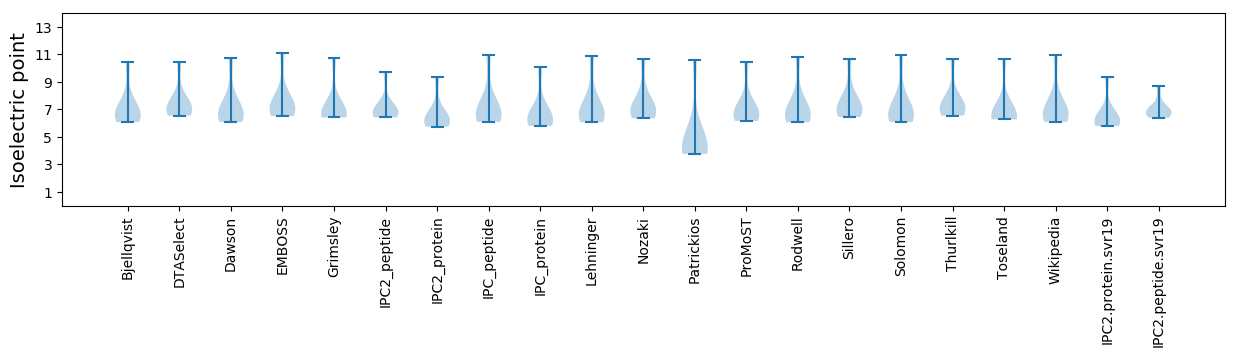
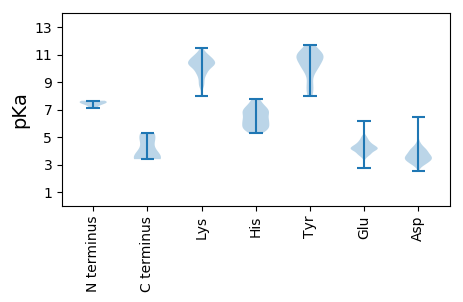
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>sp|P03132|REP68_AAV2S Protein Rep68 OS=Adeno-associated virus 2 (isolate Srivastava/1982) OX=648242 GN=Rep68 PE=1 SV=1
MM1 pKa = 7.12EE2 pKa = 4.72TQTQYY7 pKa = 10.29LTPSLSDD14 pKa = 3.2SHH16 pKa = 5.79QQPPLVWEE24 pKa = 4.86LIRR27 pKa = 11.84WLQAVAHH34 pKa = 5.29QWQTITRR41 pKa = 11.84APTEE45 pKa = 3.82WVIPRR50 pKa = 11.84EE51 pKa = 3.73IGIAIPHH58 pKa = 6.11GWATEE63 pKa = 4.09SSPPAPEE70 pKa = 5.12PGPCPPTTTTSTNKK84 pKa = 9.57FPANQEE90 pKa = 3.62PRR92 pKa = 11.84TTITTLATAPLGGILTSTDD111 pKa = 2.83STATFHH117 pKa = 7.0HH118 pKa = 6.19VTGKK122 pKa = 10.44DD123 pKa = 3.15SSTTTGDD130 pKa = 3.14SDD132 pKa = 4.04PRR134 pKa = 11.84DD135 pKa = 3.79STSSSLTFKK144 pKa = 10.5SKK146 pKa = 10.04RR147 pKa = 11.84SRR149 pKa = 11.84RR150 pKa = 11.84MTVRR154 pKa = 11.84RR155 pKa = 11.84RR156 pKa = 11.84LPITLPARR164 pKa = 11.84FRR166 pKa = 11.84CLLTRR171 pKa = 11.84STSSRR176 pKa = 11.84TSSARR181 pKa = 11.84RR182 pKa = 11.84IKK184 pKa = 10.47DD185 pKa = 3.01ASRR188 pKa = 11.84RR189 pKa = 11.84SQQTSSWCHH198 pKa = 6.8SMDD201 pKa = 3.28TSPP204 pKa = 5.28
MM1 pKa = 7.12EE2 pKa = 4.72TQTQYY7 pKa = 10.29LTPSLSDD14 pKa = 3.2SHH16 pKa = 5.79QQPPLVWEE24 pKa = 4.86LIRR27 pKa = 11.84WLQAVAHH34 pKa = 5.29QWQTITRR41 pKa = 11.84APTEE45 pKa = 3.82WVIPRR50 pKa = 11.84EE51 pKa = 3.73IGIAIPHH58 pKa = 6.11GWATEE63 pKa = 4.09SSPPAPEE70 pKa = 5.12PGPCPPTTTTSTNKK84 pKa = 9.57FPANQEE90 pKa = 3.62PRR92 pKa = 11.84TTITTLATAPLGGILTSTDD111 pKa = 2.83STATFHH117 pKa = 7.0HH118 pKa = 6.19VTGKK122 pKa = 10.44DD123 pKa = 3.15SSTTTGDD130 pKa = 3.14SDD132 pKa = 4.04PRR134 pKa = 11.84DD135 pKa = 3.79STSSSLTFKK144 pKa = 10.5SKK146 pKa = 10.04RR147 pKa = 11.84SRR149 pKa = 11.84RR150 pKa = 11.84MTVRR154 pKa = 11.84RR155 pKa = 11.84RR156 pKa = 11.84LPITLPARR164 pKa = 11.84FRR166 pKa = 11.84CLLTRR171 pKa = 11.84STSSRR176 pKa = 11.84TSSARR181 pKa = 11.84RR182 pKa = 11.84IKK184 pKa = 10.47DD185 pKa = 3.01ASRR188 pKa = 11.84RR189 pKa = 11.84SQQTSSWCHH198 pKa = 6.8SMDD201 pKa = 3.28TSPP204 pKa = 5.28
Molecular weight: 22.6 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
3936 |
204 |
735 |
492.0 |
55.28 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.479 ± 0.414 | 1.474 ± 0.353 |
5.589 ± 0.231 | 5.208 ± 0.542 |
4.395 ± 0.18 | 6.428 ± 0.585 |
2.363 ± 0.09 | 4.268 ± 0.431 |
5.716 ± 0.803 | 6.174 ± 0.255 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.956 ± 0.126 | 5.996 ± 0.553 |
6.225 ± 0.562 | 5.488 ± 0.178 |
4.751 ± 0.345 | 8.054 ± 0.621 |
7.342 ± 0.871 | 6.352 ± 0.555 |
2.261 ± 0.098 | 3.481 ± 0.376 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
